In this post, I will present a classification job on an unbalanced dataset. In an unbalanced dataset, the target variable has an uneven distribution of observations. I will be using tidymodels for the prediction workflow, BAdatasets to retrieve the data and themis to perform undersampling.
library(tidymodels)
library(themis)
library(BAdatasets)We will be using the LoanDefaults dataset. It is a dataset of defaults payments in Taiwan, presented in Yeh & Lien (2009).
data("LoanDefaults")When plotting the proportion of observations for each case, we observe that the number of negative cases (not defaulted) is much larger than the positive cases (defaulted).
LoanDefaults %>%
ggplot(aes(factor(default))) +
geom_bar(aes(y = (..count..)/sum(..count..))) +
scale_y_continuous(labels=percent) +
labs(x = "default", y = "% of cases") +
theme_minimal()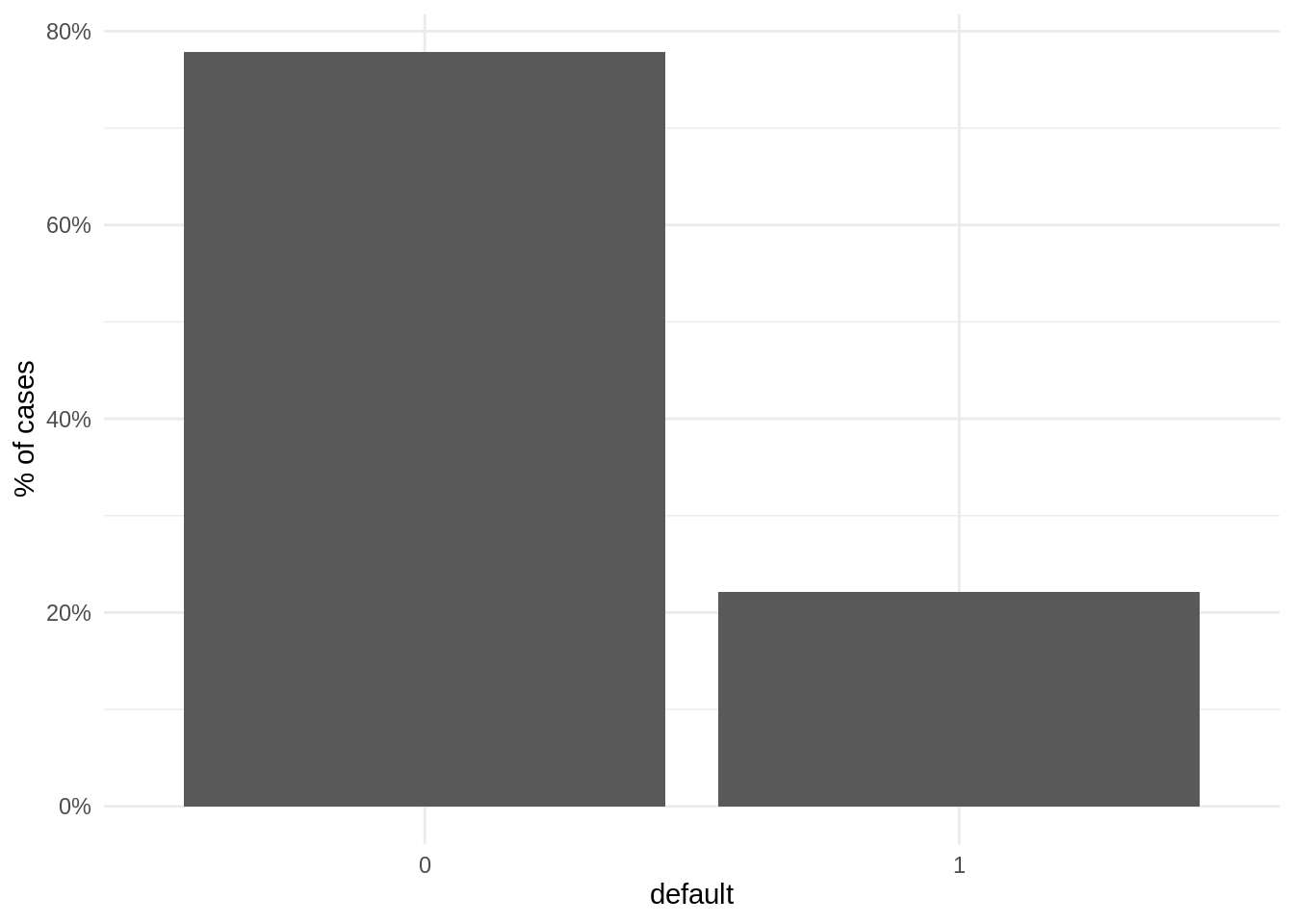
Setting workflow elements
Let’s define the elements of the tidymodels workflow. We start transforming the target default variable into a factor, setting the level of positive cases first.
LoanDefaults <- LoanDefaults %>%
mutate(default = factor(default, levels = c("1", "0")))Then, we need to split the data into train and test. As the dataset is large, I have selected a large prop value. Being the dataset unbalanced, it is important to set strata = default, so that the distribution of target variable in train and test sets will be similar to the original dataset. I have applied a similar logic to split the training set into four folds to apply cross validation.
set.seed(1313)
split <- initial_split(LoanDefaults, prop = 0.9, strata = "default")
folds <- vfold_cv(training(split), v = 10, strata = "default")To evaluate model performance, I have chosen the following metrics:
accuracy: fraction of observations correctly classified.- sensitivity
sens: the fraction of positive elements correctly classified. - specificity
spec: the fraction of negative elements correctly classified. - area under the ROC curve
roc_auc: a parameter assessing the tradeoff betweensensandspec.
class_metrics <- metric_set(accuracy, sens, spec, roc_auc)I will be using two models to train: a decision tree dt and a random forest rf. I will set standard values of parameter for each model. Hyperparameter tuning (not presented here) yields little influence of these parameters in model results.
dt <- decision_tree(mode = "classification", cost_complexity = 0.1, min_n = 10) %>%
set_engine("rpart")
rf <- rand_forest(mode = "classification", trees = 50, mtry = 5, min_n = 10) %>%
set_engine("ranger")The preprocessing recipe is quite complex in this case:
- the variable
IDis removed. - variables
PAY0toPAY6are removed. Before that, apayment_statusvariable is defined fromPAY_0. - variables
payment_status,MARRIAGEandEDUCATIONare transformed so that abormal values are set toNA. NAvalues of predictors are imputed usign k nearest neighbors.SEX,MARRIAGEandEDUCATIONare transformed into factors.
rec_unb <- training(split) %>%
recipe(default ~ .) %>%
step_rm(ID) %>%
step_mutate(payment_status = ifelse(PAY_0 < 1, 0, PAY_0)) %>%
step_rm(PAY_0:PAY_6) %>%
step_num2factor(SEX, levels = c("male", "female")) %>%
step_mutate(MARRIAGE = ifelse(!MARRIAGE %in% 1:3, NA, MARRIAGE)) %>%
step_mutate(EDUCATION = ifelse(!EDUCATION %in% 1:4, NA, EDUCATION)) %>%
step_num2factor(MARRIAGE, levels = c("marriage", "single", "others")) %>%
step_num2factor(EDUCATION, levels = c("graduate", "university", "high_school", "others")) %>%
step_impute_knn(all_predictors(), neighbors = 3)Now we can test the two models using cross validation. We start with the decision tree:
dt_cv_unb <- fit_resamples(object = dt,
preprocessor = rec_unb,
resamples = folds,
metrics = class_metrics)
rf_cv_unb <- fit_resamples(object = rf,
preprocessor = rec_unb,
resamples = folds,
metrics = class_metrics)The metrics for the decision tree are:
dt_cv_unb %>%
collect_metrics()## # A tibble: 4 × 6
## .metric .estimator mean n std_err .config
## <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 accuracy binary 0.820 10 0.00140 Preprocessor1_Model1
## 2 roc_auc binary 0.644 10 0.00321 Preprocessor1_Model1
## 3 sens binary 0.329 10 0.00661 Preprocessor1_Model1
## 4 spec binary 0.959 10 0.000712 Preprocessor1_Model1And the metrics for cross validation:
rf_cv_unb %>%
collect_metrics()## # A tibble: 4 × 6
## .metric .estimator mean n std_err .config
## <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 accuracy binary 0.818 10 0.00242 Preprocessor1_Model1
## 2 roc_auc binary 0.759 10 0.00374 Preprocessor1_Model1
## 3 sens binary 0.360 10 0.00680 Preprocessor1_Model1
## 4 spec binary 0.948 10 0.00176 Preprocessor1_Model1The sensitivity of the two models is quite low. This is not good, because it means that a default will be undetected using these models.
Model with undersampling
A commonly used technique to improve classification of unbalanced data is to modify the training set so that it has the same number of positives and negatives. There are two ways of doing this:
- oversampling: creating additional artificial observations for the less frequent case.
- undersampling: removing observations of the most frequent case.
In tidymodels, those recipes are implemented with the themis package.
For this dataset, I have decided to undersample the training set using the step_downsample. The use of oversampling has lead to unsatisfactory results.
rec_us <- rec_unb %>%
step_downsample(default, under_ratio = 1)Let’s examine the distribution of the target variable after downsampling:
rec_us %>%
prep() %>%
juice() %>%
ggplot(aes(factor(default))) +
geom_bar(aes(y = (..count..)/sum(..count..))) +
scale_y_continuous(labels=percent) +
labs(x = "default", y = "% of cases") +
theme_minimal()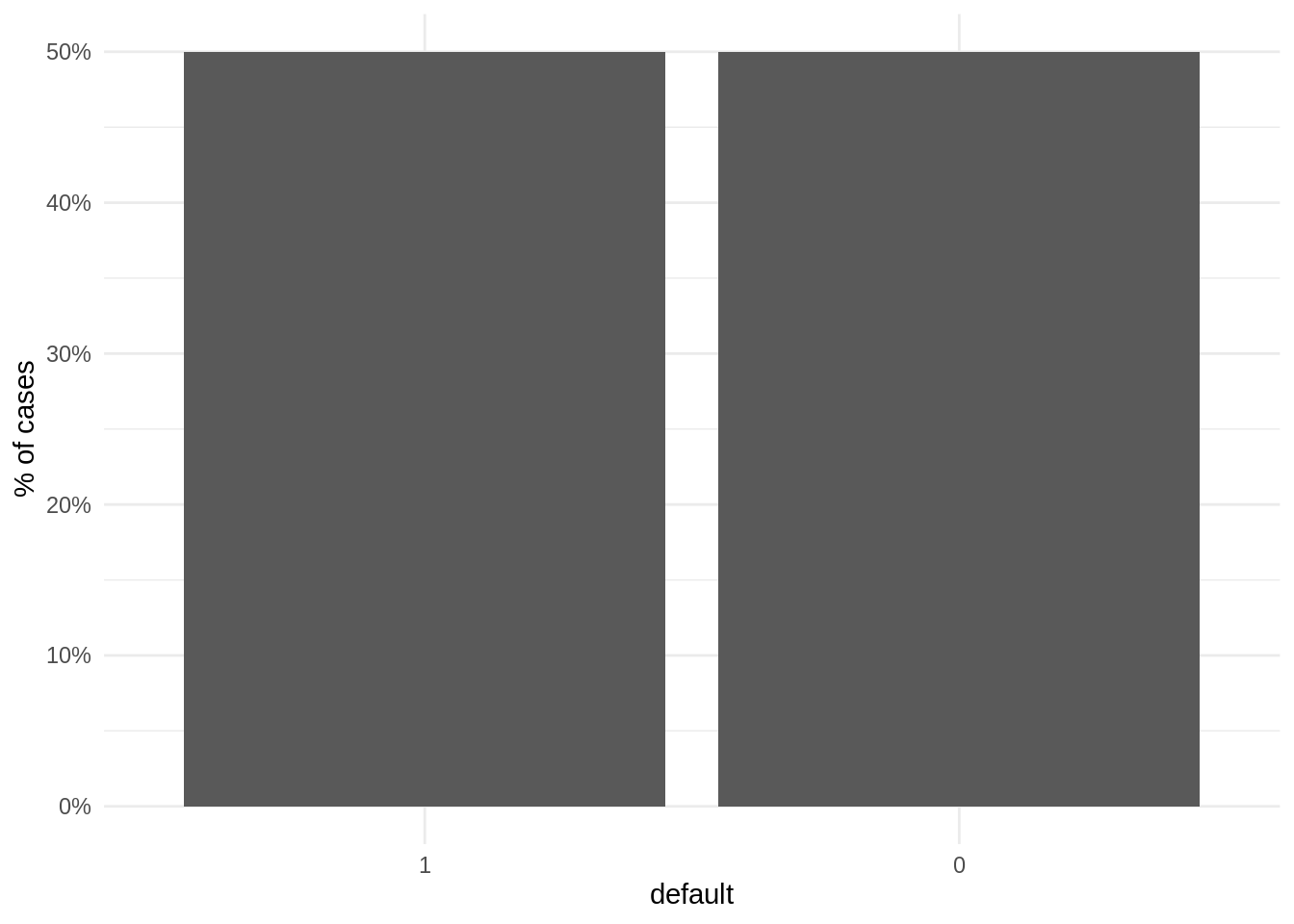
We observe that all the dataset for which is trained the model is balanced. Let’s proceed to train the models with the undersampled recipe:
dt_cv_us <- fit_resamples(object = dt,
preprocessor = rec_us,
resamples = folds,
metrics = class_metrics)
rf_cv_us <- fit_resamples(object = rf,
preprocessor = rec_us,
resamples = folds,
metrics = class_metrics)As the metrics are presented in data frames, we can put them all together and examine the performance of the four models at once. As variability of metrics can be an issue when balancing data, I have added errorbars for each parameter.
t1 <- dt_cv_unb %>%
collect_metrics() %>%
mutate(model = "tree", train = "unbalanced")
t2 <- rf_cv_unb %>%
collect_metrics() %>%
mutate(model = "forest", train = "unbalanced")
t3 <- dt_cv_us %>%
collect_metrics() %>%
mutate(model = "tree", train = "balanced")
t4 <- rf_cv_us %>%
collect_metrics() %>%
mutate(model = "forest", train = "balanced")
bind_rows(t1, t2, t3, t4) %>%
ggplot(aes(x =.metric, y = mean, ymin = mean - std_err, ymax = mean + std_err, fill = train)) +
geom_bar(stat = "identity", position = "dodge") +
geom_errorbar(position=position_dodge(width=1), width = 0.5) +
scale_fill_manual(values = c("#990000", "#FF8000")) +
facet_grid(. ~ model) +
labs(x = "metric", y = "value")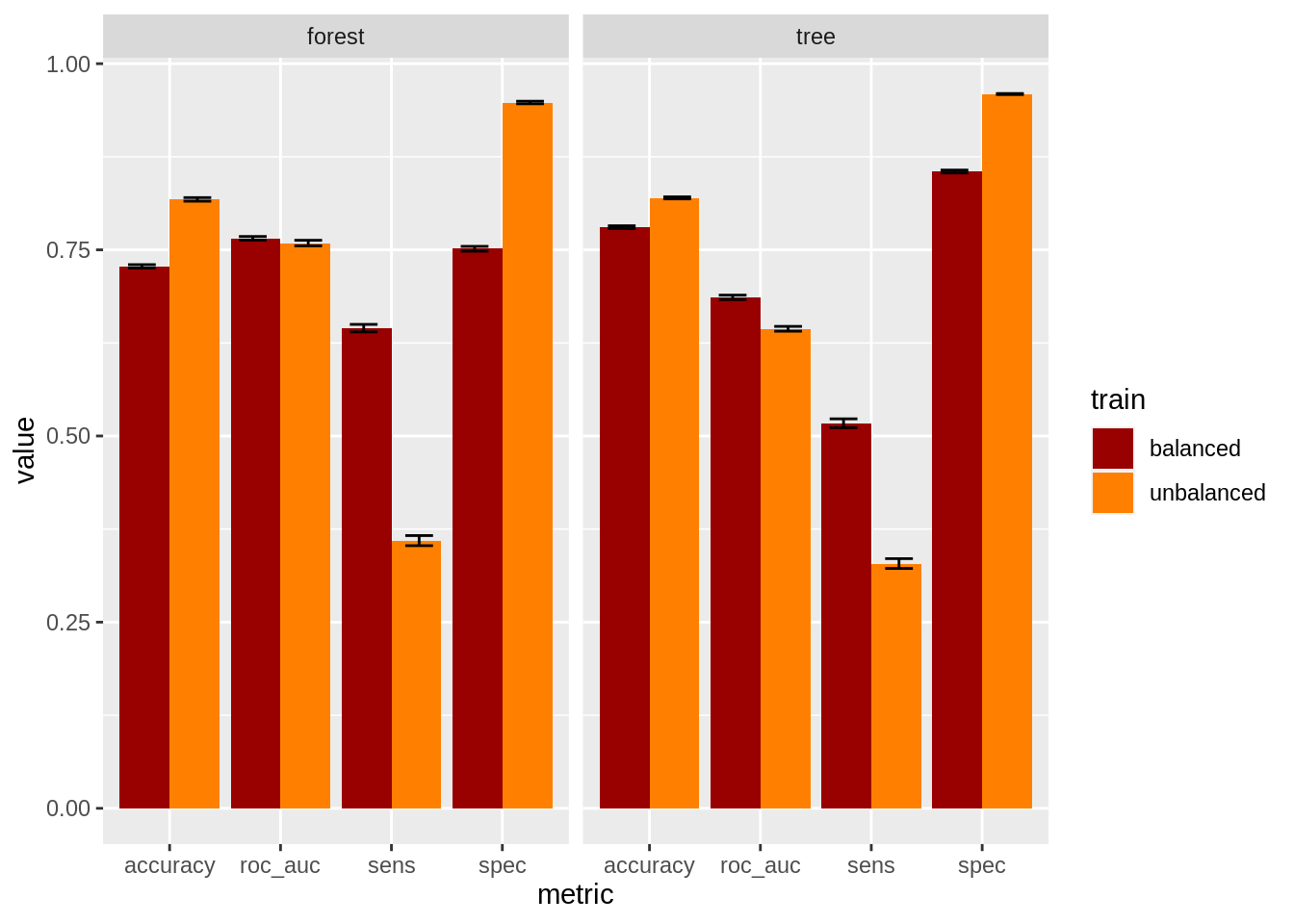
We can observe that undersampling increases values of sensitivity at the cost of reducing specificity. Undersampling leads to slightly worse values of accuracy, and slightly better values of AUC.
Prediction for the undersampled model
Let’s choose the undersampled model trained with the random forest model. We can test this model on both the test and train set.
We start defining the model using a workflow:
rf_us <- workflow() %>%
add_recipe(rec_us) %>%
add_model(rf) %>%
fit(training(split))Let’s obtain the predicted values for train and test sets. Note that the prediction on the train set is with the original data, not with the undersampled ones. Undersampling only takes place to fit the model.
pred_train <- rf_us %>%
predict(training(split)) %>%
bind_cols(training(split) %>% select(default))
pred_test <- rf_us %>%
predict(testing(split)) %>%
bind_cols(testing(split) %>% select(default))The confusion matrices show that the model tends to inflate false positives to obtain a decent value of specifity:
class_metrics2 <- metric_set(accuracy, sens, spec)
pred_train %>%
conf_mat(truth = default, estimate = .pred_class)## Truth
## Prediction 1 0
## 1 5870 3941
## 0 102 17086pred_test %>%
conf_mat(truth = default, estimate = .pred_class)## Truth
## Prediction 1 0
## 1 430 572
## 0 234 1765Let’s compare accuracy, sensitivity and specificity for train and test sets:
class_metrics2 <- metric_set(accuracy, sens, spec)
t_train <- pred_train %>%
class_metrics2(truth = default, estimate = .pred_class) %>%
mutate(set = "train")
t_test <- pred_test %>%
class_metrics2(truth = default, estimate = .pred_class) %>%
mutate(set = "test")
bind_rows(t_train, t_test) %>%
ggplot(aes(.metric, .estimate, fill = set)) +
geom_bar(stat = "identity", position = "dodge") +
scale_fill_manual(values = c("#990000", "#FF8000")) +
labs(x = "metric", y = "value") +
theme_minimal()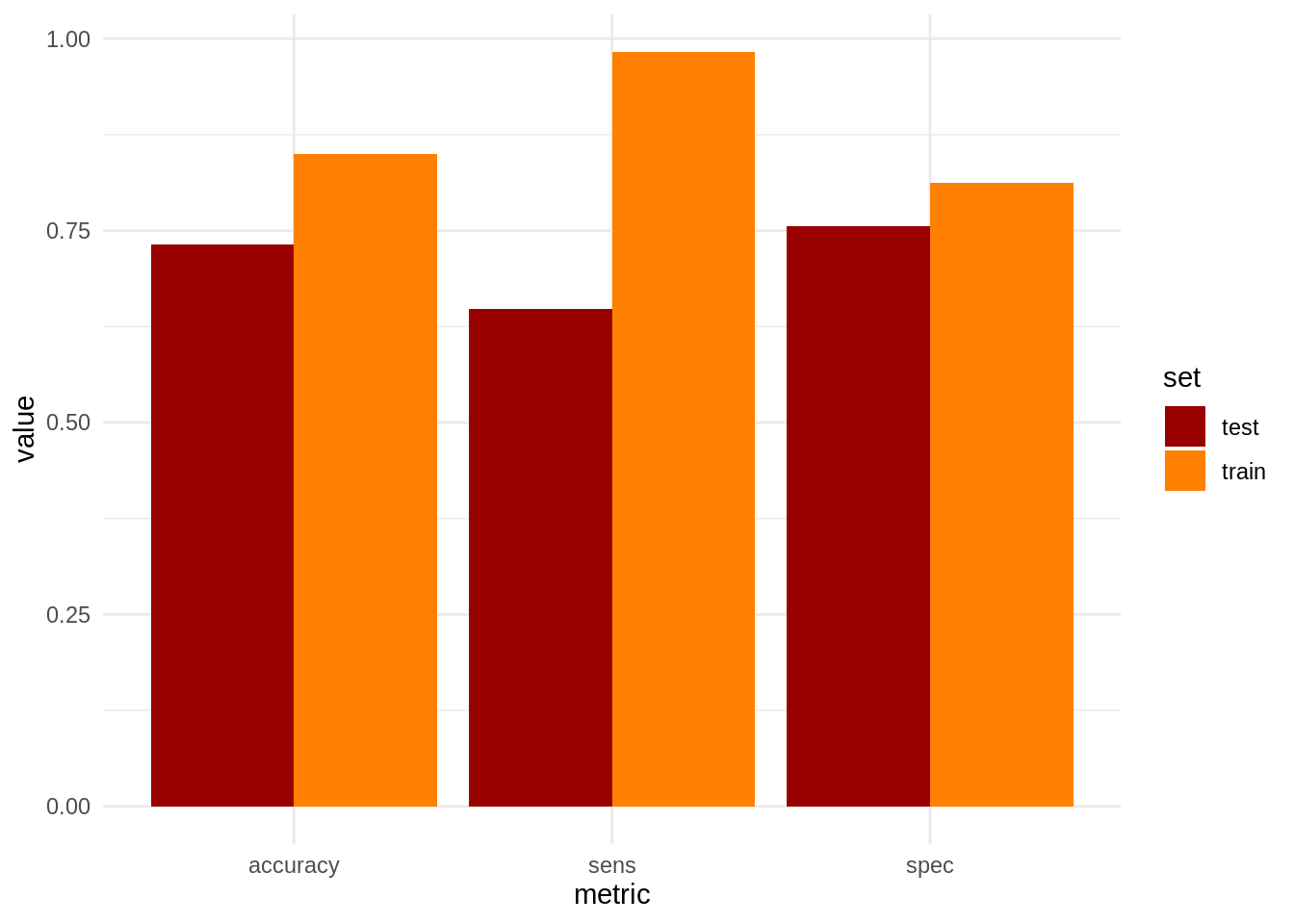
We observe that undersampling has improved sensitivity at the price of worsening accuracy and specificity. In this case, undersampling has somewhat improved our model, but this is not always the case. Specially when we use oversampling, where we introduce artificial observations to train the model. Some alternative approaches to deal with unbalanced datasets can be found at the StackExchange question cited below.
References
- BAdatasets page: https://github.com/jmsallan/BAdatasets
themispackage website: https://themis.tidymodels.org/- StackExchange question: Are unbalanced datasets problematic, and (how) does oversampling (purport to) help? https://stats.stackexchange.com/questions/357466/are-unbalanced-datasets-problematic-and-how-does-oversampling-purport-to-he
- Yeh, I. C., & Lien, C. H. (2009). The comparisons of data mining techniques for the predictive accuracy of probability of default of credit card clients. Expert Systems with Applications, 36(2), 2473-2480. https://doi.org/10.1016/j.eswa.2007.12.020
Session info
sessionInfo()## R version 4.2.1 (2022-06-23)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Linux Mint 19.2
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/openblas/libblas.so.3
## LAPACK: /usr/lib/x86_64-linux-gnu/libopenblasp-r0.2.20.so
##
## locale:
## [1] LC_CTYPE=es_ES.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=es_ES.UTF-8 LC_COLLATE=es_ES.UTF-8
## [5] LC_MONETARY=es_ES.UTF-8 LC_MESSAGES=es_ES.UTF-8
## [7] LC_PAPER=es_ES.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=es_ES.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] ranger_0.13.1 rpart_4.1.16 BAdatasets_0.1.0 themis_0.2.1
## [5] yardstick_0.0.9 workflowsets_0.2.1 workflows_0.2.6 tune_0.2.0
## [9] tidyr_1.2.0 tibble_3.1.6 rsample_0.1.1 recipes_0.2.0
## [13] purrr_0.3.4 parsnip_0.2.1 modeldata_0.1.1 infer_1.0.0
## [17] ggplot2_3.3.5 dplyr_1.0.9 dials_0.1.1 scales_1.2.0
## [21] broom_0.8.0 tidymodels_0.2.0
##
## loaded via a namespace (and not attached):
## [1] lubridate_1.8.0 doParallel_1.0.17 DiceDesign_1.9 tools_4.2.1
## [5] backports_1.4.1 bslib_0.3.1 utf8_1.2.2 R6_2.5.1
## [9] DBI_1.1.2 colorspace_2.0-3 nnet_7.3-17 withr_2.5.0
## [13] tidyselect_1.1.2 parallelMap_1.5.1 compiler_4.2.1 cli_3.3.0
## [17] labeling_0.4.2 bookdown_0.26 sass_0.4.1 checkmate_2.1.0
## [21] stringr_1.4.0 digest_0.6.29 rmarkdown_2.14 unbalanced_2.0
## [25] pkgconfig_2.0.3 htmltools_0.5.2 parallelly_1.31.1 lhs_1.1.5
## [29] highr_0.9 fastmap_1.1.0 rlang_1.0.2 rstudioapi_0.13
## [33] BBmisc_1.12 FNN_1.1.3 farver_2.1.0 jquerylib_0.1.4
## [37] generics_0.1.2 jsonlite_1.8.0 magrittr_2.0.3 ROSE_0.0-4
## [41] Matrix_1.5-1 Rcpp_1.0.8.3 munsell_0.5.0 fansi_1.0.3
## [45] GPfit_1.0-8 lifecycle_1.0.1 furrr_0.3.0 stringi_1.7.6
## [49] pROC_1.18.0 yaml_2.3.5 MASS_7.3-58 plyr_1.8.7
## [53] grid_4.2.1 parallel_4.2.1 listenv_0.8.0 crayon_1.5.1
## [57] lattice_0.20-45 splines_4.2.1 mlr_2.19.0 knitr_1.39
## [61] pillar_1.7.0 future.apply_1.9.0 codetools_0.2-18 fastmatch_1.1-3
## [65] glue_1.6.2 evaluate_0.15 ParamHelpers_1.14 blogdown_1.9
## [69] data.table_1.14.2 vctrs_0.4.1 foreach_1.5.2 RANN_2.6.1
## [73] gtable_0.3.0 future_1.25.0 assertthat_0.2.1 xfun_0.30
## [77] gower_1.0.0 prodlim_2019.11.13 class_7.3-20 survival_3.4-0
## [81] timeDate_3043.102 iterators_1.0.14 hardhat_0.2.0 lava_1.6.10
## [85] globals_0.14.0 ellipsis_0.3.2 ipred_0.9-12