In this post, I will introduce regularized regression, and then use the glmnet package to evaluate a regularized regression model on the InsuranceCharges dataset.
InsuranceCharges contains several features of individuals such as age, physical/family condition and location, and their existing medical expense. We intend to predict future medical expenses of individuals that help medical insurance to make decision on charging the premium. Those expenses are in the charges variable.
The data are embedded in the BAdatasets package. I’ll load also recipes for data preprocessing, yardstick for performance metrics and glmnet for regularized regression.
library(recipes)
library(yardstick)
library(BAdatasets)
library(glmnet)I have performed some transformations of the dataset using the recipes package:
- replace the continuous variable
bmiby abmi30dummy variable, splitting the data into individuals with BMI less or equal than 30 and larger than 30. - adding a quadratic term to
age. - transform all factors (nominal variables) into dummies using one hot encoding and then remove one of the dummies for each category.
- add an interaction term between
bmi30andsmoker_yes.
rec <- InsuranceCharges %>%
recipe(charges ~ .) %>%
step_mutate(bmi30 = ifelse(bmi <= 30, 0 , 1)) %>%
step_rm(bmi) %>%
step_poly(age, degree = 2) %>%
step_dummy(all_nominal(), one_hot = TRUE) %>%
step_rm(sex_male, smoker_no, region_northeast) %>%
step_interact(terms = ~ bmi30:smoker_yes)The transformed data are in the insurance data frame:
insurance <- rec %>%
prep() %>%
juice()The result is a new dataset with 11 predictors:
insurance %>%
glimpse()## Rows: 1,338
## Columns: 11
## $ children <int> 0, 1, 3, 0, 0, 0, 1, 3, 2, 0, 0, 0, 0, 0, 0, 1, 1, …
## $ charges <dbl> 16884.924, 1725.552, 4449.462, 21984.471, 3866.855,…
## $ bmi30 <dbl> 0, 1, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 1, 1, 0, 1, …
## $ age_poly_1 <dbl> -0.039333409, -0.041279930, -0.021814716, -0.012082…
## $ age_poly_2 <dbl> 0.036256381, 0.043000157, -0.010053464, -0.02459349…
## $ sex_female <dbl> 1, 0, 0, 0, 0, 1, 1, 1, 0, 1, 0, 1, 0, 1, 0, 0, 1, …
## $ smoker_yes <dbl> 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 0, …
## $ region_northwest <dbl> 0, 0, 0, 1, 1, 0, 0, 1, 0, 1, 0, 0, 0, 0, 0, 0, 0, …
## $ region_southeast <dbl> 0, 1, 1, 0, 0, 1, 1, 0, 0, 0, 0, 1, 0, 1, 1, 0, 0, …
## $ region_southwest <dbl> 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, …
## $ bmi30_x_smoker_yes <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, …Regularized regression
In datasets with many features, it can happen that using all of them leads to a worse performance than using only a subset. Selecting the features to introduce for better accuracy is the feature selection problem. There are two classical techniques of feature selection:
- Best subsets, which examines the fit of all possible subsets of features to choose the best subset. This can be computationally expensive. Sometimes we use algorithms like simulated annealing to choose that subset.
- Stepwise regression, consisting in adding or removing a feature in each step, until a satisfactory model is found.
An alternative to these strategies is regularization, in which we limit the total value of regression coefficients, so the regression coefficients of some variables shrink or go to zero.
The regression coefficients in the ordinary least squares (OLS) approach to linear regression are such that minimize the sum of squared errors:
\[ \displaystyle\sum_{i=1}^n \left( y_i - \hat{y}_i \right)^2 \]
Let’s examine two regularization approaches to OLS, lasso and ridge regression.
The name lasso stands for least absolute shrinkage and selection operator.
The approach of lasso is to force regression coefficients to shrink, forcing that the sum of absolute values or 1-norm to be smaller than a value \(t\):
\[ \displaystyle\sum_{j=1}^p |\beta_j| = \lVert \beta \lVert_1 \leq t\]
This is equivalent to minimizing:
\[ \displaystyle\sum_{i=1}^n \left( y_i - \hat{y}_i \right)^2 + \lambda \lVert \beta \lVert_1 \]
Lasso tends to set to zero some regression coefficients, so it works as an automated feature selection tool. As it uses the 1-norm of regression coefficients, lasso is also called L1 regularization.
Ridge regression uses a strategy analogous to lasso regression, but bounding the 2-norm (sum of squares) of the vector of regression coefficients to a value \(t\):
\[ \sqrt{\displaystyle\sum_{j=1}^p \beta_j^2} = \lVert \beta \lVert_2 \leq t \]
This is equivalent to minimizing:
\[ \displaystyle\sum_{i=1}^n \left( y_i - \hat{y}_i \right)^2 + \lambda \lVert \beta \lVert_2 \]
As ridge regression uses the 2-norm, we call it L2 regularization.
Ridge regression improves prediction error by shrinking coefficients of some variables.
We can mixt both regularizations in a mixed model introducing a mixture parameter which ranges from \(\alpha = 0\) for ridge regression and to \(\alpha = 1\) for lasso regression. \(\alpha\) is sometimes called the mixture parameter. These models are sometimes called elastic nets.
\[ \displaystyle\sum_{i=1}^n \left( y_i - \hat{y}_i \right)^2 + \lambda\left( \alpha \lVert \beta \lVert_2 + \left(1 - \alpha \right) \lVert \beta \lVert_1 \right) \]
The results of all these models depend on a \(\lambda\) parameter. As regularized regression is used mainly for prediction, we choose the value of lambda that best fits a prediction metric, usually the mean squared error. Let’s see how we can do that using glmnet.
A ridge regression model
glmnet requires entering variables in matrix format, that’s why I have generated dummies for categorical variables. I obtain the inputs to glmnet doing:
dep <- insurance %>%
pull(charges)
vars <- insurance %>%
select(-charges) %>%
as.matrix()Let’s obtain the ridge model:
ridge <- glmnet(x = vars, y = dep, alpha = 0)glmnet has picked a range of values of \(\lambda\) and obtained the model for each. Let’s plot the evolution of coefficients with \(\lambda\):
plot(ridge, xvar = "lambda")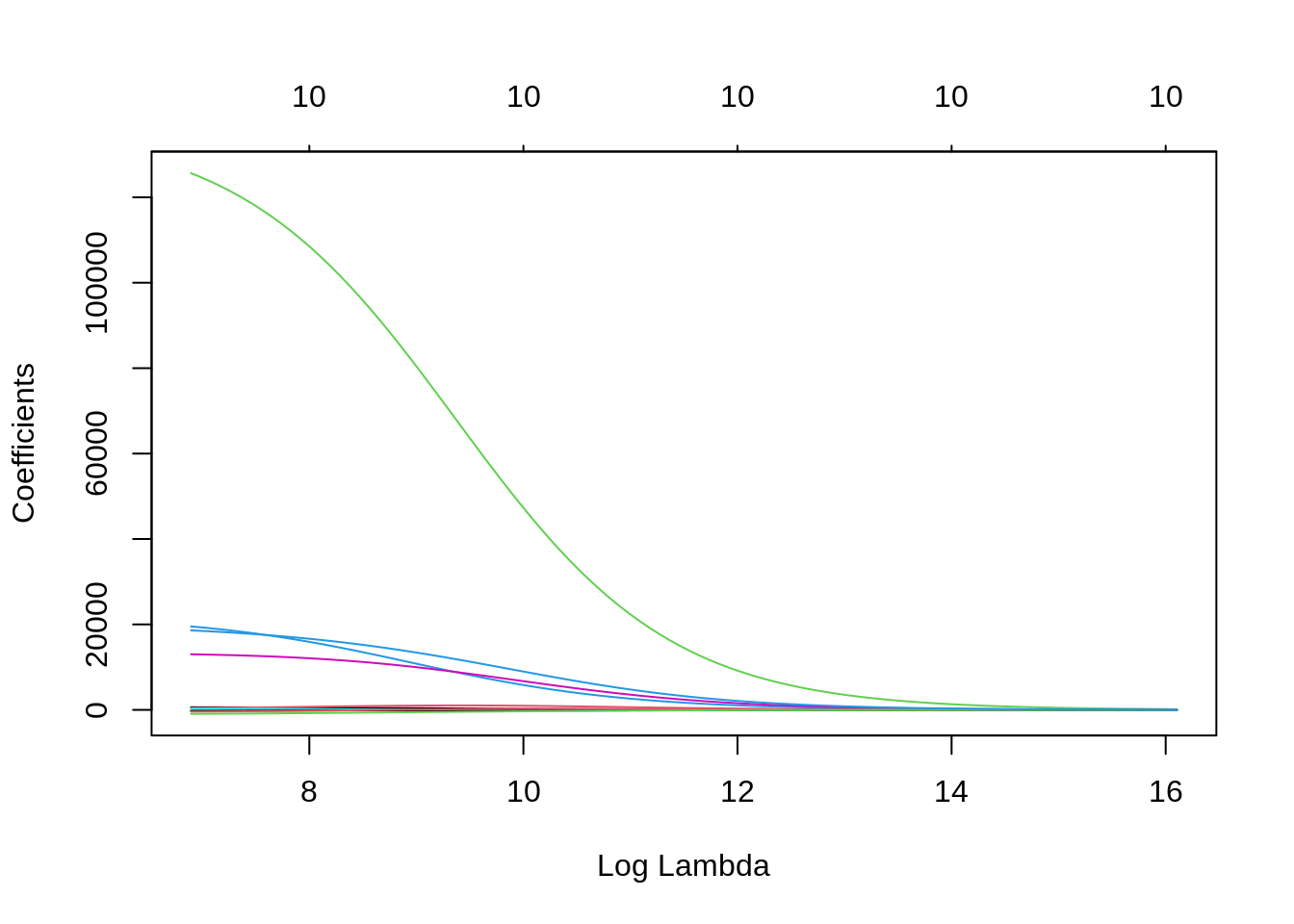
For high values of \(\lambda\), the coefficients of all variables are shrinking. Small values of \(\lambda\) result in coefficients similar to OLS. We need to select the value of \(\lambda\) for our prediction job as the one that minimizes a performance metric for prediction. cv.glmnet does that using cross validation.
cv_ridge <- cv.glmnet(x = vars,
y = dep,
alpha = 0)Here are the results as a function of \(\lambda\):
plot(cv_ridge)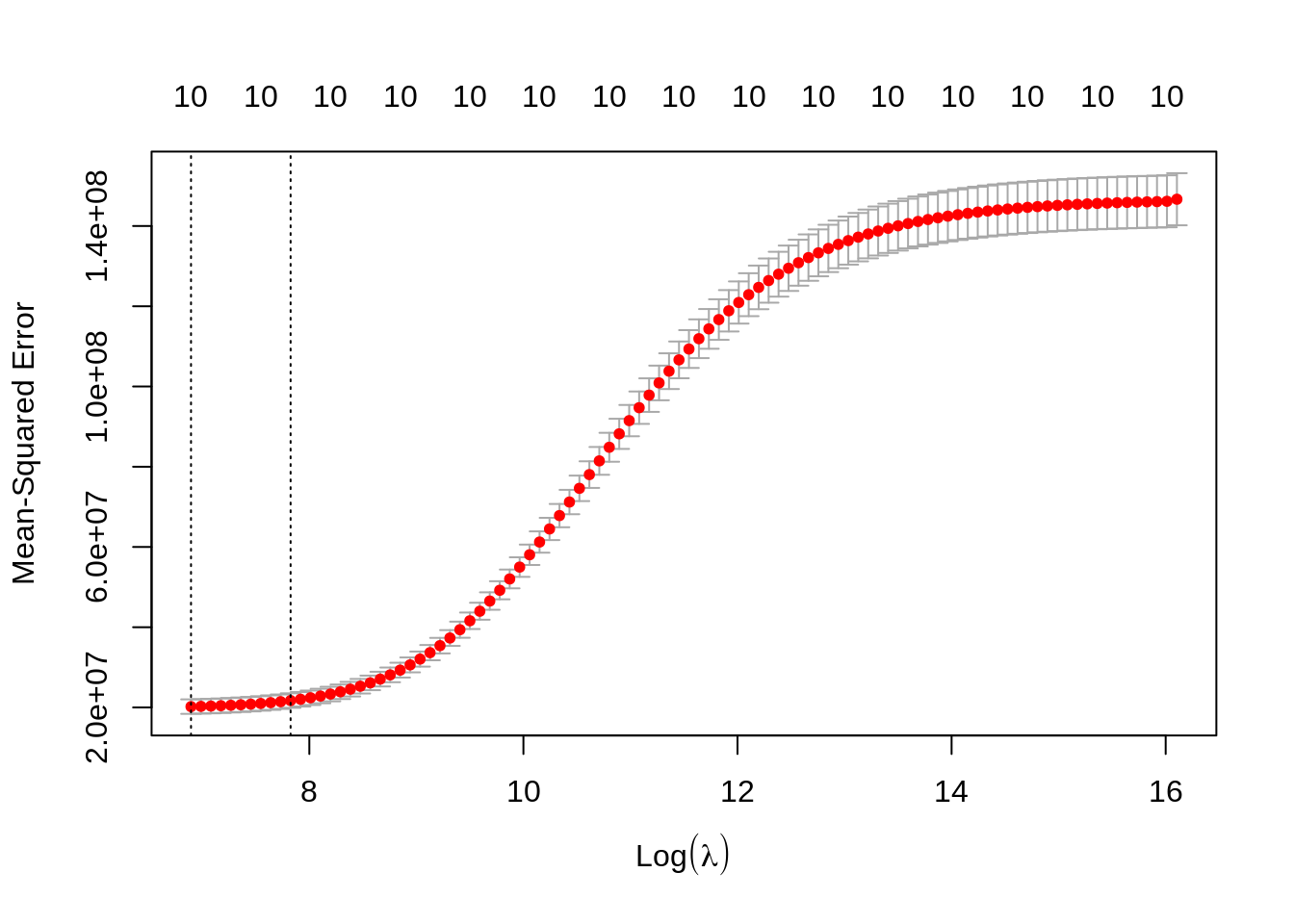
We get two values of \(\lambda\): the optimum lambda.min and lambda.1se which gives a similar fit with some more shrinking of coefficients. We obtain the values predicted with lambda.min doing:
pred_lambdamin_ridge <- predict(cv_ridge,
s = cv_ridge$lambda.min,
newx = vars)[, 1]Lasso regression
To fit a lasso regression we set alpha = 1:
lasso <- glmnet(x = vars, y = dep, alpha = 1)Let’s plot the evolution of coefficients:
plot(lasso, xvar = "lambda")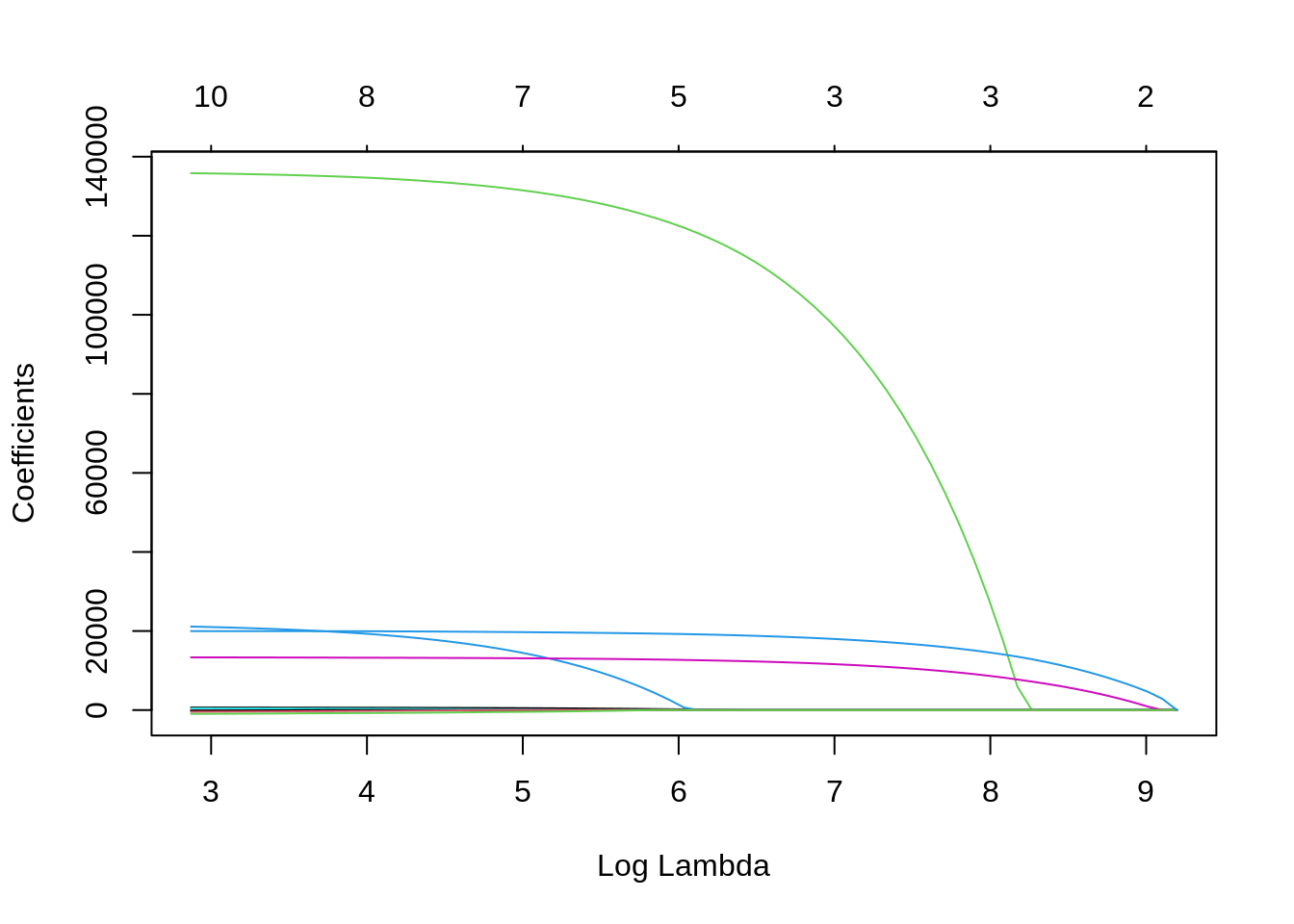
Lasso sets to zero coefficients of predictors as \(\lambda\) increases, so it can be considered an automated feature selector. The number of variables selected for each \(\lambda\) are presented in the scale above the plot. Let’s select \(\lambda\) with cross validation and plot the result:
cv_lasso <- cv.glmnet(x = vars,
y = dep,
alpha = 1,
penalty.factor = rep(1, 10))
plot(cv_lasso)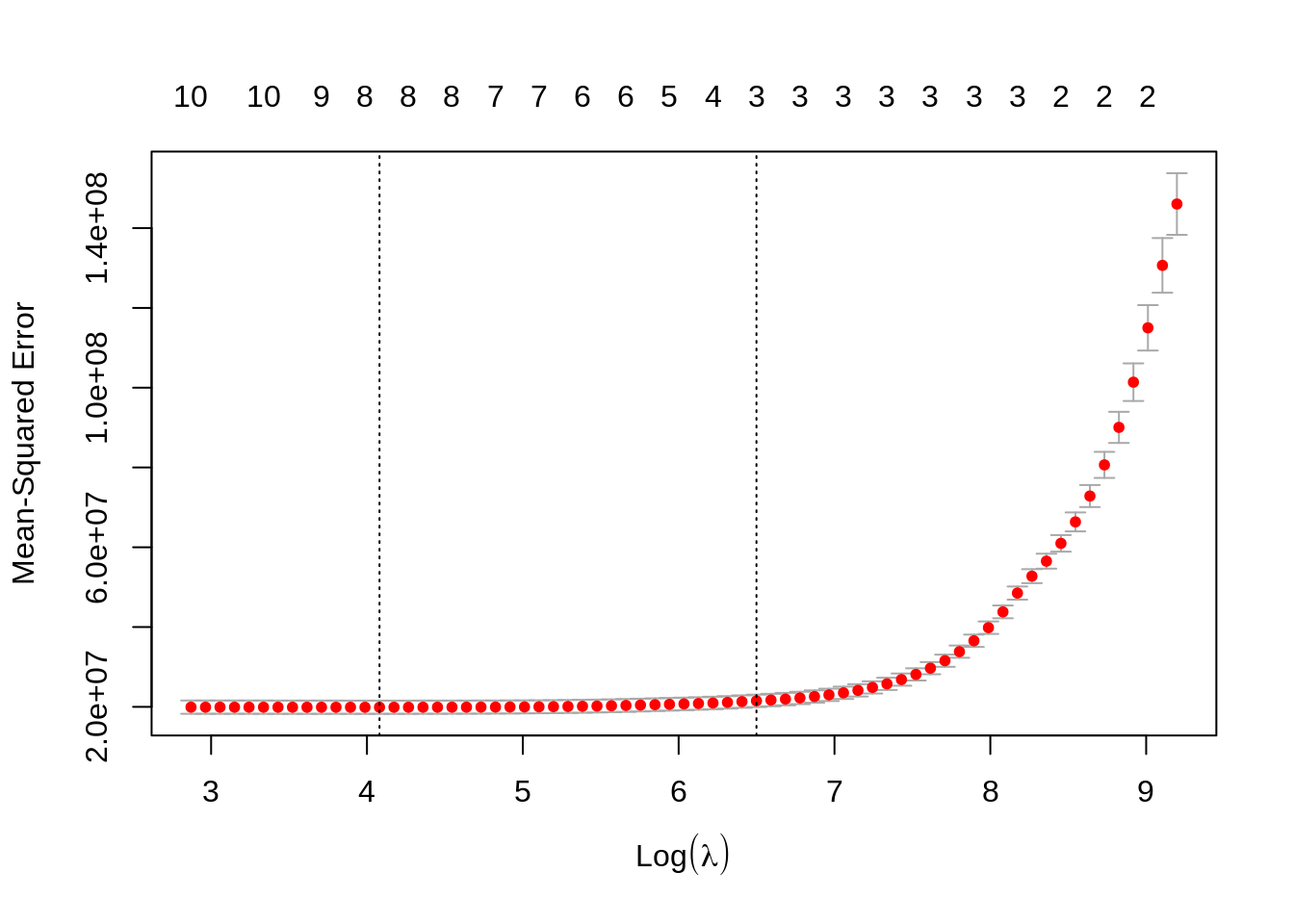
Finally, let’s do the prediction using lambda.min like in ridge regression.
pred_lambdamin_lasso <- predict(cv_lasso,
s = cv_lasso$lambda.min,
newx = vars)[, 1]Model performance
I will use yardstick to evaluate performance of lasso and ridge regression. Let’s define a metric_set for numerical prediction.
np_metrics <- metric_set(rmse, mae, rsq)Let’s store observed values and predictions in a data frame.
np_metrics <- metric_set(rmse, mae, rsq)
predictions <- tibble(real = dep,
ridge = pred_lambdamin_ridge,
lasso = pred_lambdamin_lasso) The performance of ridge regression:
predictions %>%
np_metrics(truth = real, estimate = ridge)## # A tibble: 3 × 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 rmse standard 4458.
## 2 mae standard 2574.
## 3 rsq standard 0.867And the performance of lasso regression:
predictions %>%
np_metrics(truth = real, estimate = lasso)## # A tibble: 3 × 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 rmse standard 4423.
## 2 mae standard 2467.
## 3 rsq standard 0.867Both models have quite similar performance in this case.
References
- Hastle, T., Qian, J., Tay, K. (2021). An introduction to glmnet. https://glmnet.stanford.edu/articles/glmnet.html
- Kaggle. Insurance premium prediction- https://www.kaggle.com/noordeen/insurance-premium-prediction
- Silge, Julia (2021). Add error for ridge regression with glmnet #431. https://github.com/tidymodels/parsnip/issues/431
- UC Business Analytics R Programming Guide. Regularized regression. https://uc-r.github.io/regularized_regression
- Yang, Y. (2021). Understand penalty.factor in glmnet. https://yuyangyy.medium.com/understand-penalty-factor-in-glmnet-9fb873f9045b
Session info
## R version 4.2.0 (2022-04-22)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Linux Mint 19.2
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/openblas/libblas.so.3
## LAPACK: /usr/lib/x86_64-linux-gnu/libopenblasp-r0.2.20.so
##
## locale:
## [1] LC_CTYPE=es_ES.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=es_ES.UTF-8 LC_COLLATE=es_ES.UTF-8
## [5] LC_MONETARY=es_ES.UTF-8 LC_MESSAGES=es_ES.UTF-8
## [7] LC_PAPER=es_ES.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=es_ES.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] glmnet_4.1-4 Matrix_1.4-1 BAdatasets_0.1.0 yardstick_0.0.9
## [5] recipes_0.2.0 dplyr_1.0.9
##
## loaded via a namespace (and not attached):
## [1] Rcpp_1.0.8.3 lubridate_1.8.0 lattice_0.20-45 tidyr_1.2.0
## [5] listenv_0.8.0 class_7.3-20 assertthat_0.2.1 digest_0.6.29
## [9] ipred_0.9-12 foreach_1.5.2 utf8_1.2.2 parallelly_1.31.1
## [13] R6_2.5.1 plyr_1.8.7 hardhat_0.2.0 evaluate_0.15
## [17] highr_0.9 blogdown_1.9 pillar_1.7.0 rlang_1.0.2
## [21] rstudioapi_0.13 jquerylib_0.1.4 rpart_4.1.16 rmarkdown_2.14
## [25] splines_4.2.0 gower_1.0.0 stringr_1.4.0 compiler_4.2.0
## [29] xfun_0.30 pkgconfig_2.0.3 shape_1.4.6 globals_0.14.0
## [33] htmltools_0.5.2 nnet_7.3-17 tidyselect_1.1.2 tibble_3.1.6
## [37] prodlim_2019.11.13 bookdown_0.26 codetools_0.2-18 fansi_1.0.3
## [41] future_1.25.0 crayon_1.5.1 withr_2.5.0 MASS_7.3-57
## [45] grid_4.2.0 jsonlite_1.8.0 lifecycle_1.0.1 DBI_1.1.2
## [49] magrittr_2.0.3 pROC_1.18.0 future.apply_1.9.0 cli_3.3.0
## [53] stringi_1.7.6 timeDate_3043.102 bslib_0.3.1 ellipsis_0.3.2
## [57] generics_0.1.2 vctrs_0.4.1 lava_1.6.10 iterators_1.0.14
## [61] tools_4.2.0 glue_1.6.2 purrr_0.3.4 parallel_4.2.0
## [65] fastmap_1.1.0 survival_3.2-13 yaml_2.3.5 knitr_1.39
## [69] sass_0.4.1