Complex network analysis usually examines networks at the global level, with metrics like average path length, average clustering coefficient and global and local efficiency. They are also examined at the local level, with measures such as node degree or node and edge betweenness.
In complex networks theory, there is a stream of reseearch focusing on intermediate network structures obtained from the examination of subgraphs. A subgraph is a subset of a graph, sharing some or all of its nodes and edges. The analysis of subgraphs allows us to identify network motifs. Motifs are patterns of interconnections of a network, occurring at a number significantly higher than in randomized versions of the graph: a random graph with the same number of nodes, edges and degree distribution.
In this post, I will introduce how to use the R igraph library to identify network motifs. These network motifs can be described as isomorphism classes of subgraphs (usually between three and finve nodes) more abundant than in a random network. I will also use purrr for functional programming and data.table for tabular data handling.
library(igraph)
library(purrr)
library(data.table)Isomorphic Classes of Subgraphs
Two graphs are isomorphic when there is a matching between nodes of each graph so that two nodes connected in the first graph correspond to nodes connected in the second graph. A set of isomorphic graphs forms an isomorphic class. To be isomorphic, graphs need to have the same number of nodes, so it is frequent to consider isomorphic classes for a given number of nodes. Let’s illustrate this with two isomorphic graphs of four nodes.
g1 <- graph_from_literal(A -- B, B -- D, C -- D, C -- A)
g1 <- permute(g1, c(1, 2, 4, 3))
g2 <- graph_from_literal(A -- D, D -- B, B -- C, C -- A)
g2 <- permute(g2, c(1, 4, 2, 3))
layout4 <- matrix(c(0, 1,
1, 1,
0, 0,
1, 0), nrow = 4, byrow = TRUE)
iso4 <- list(g1, g2)
par(mfrow = c(1, 2), oma = c(0.5, 1, 0.5, 1), mar = c(1, 1, 1, 1))
walk(iso4, ~ plot(., vertex.size = 40, vertex.color = "white", layout = layout4))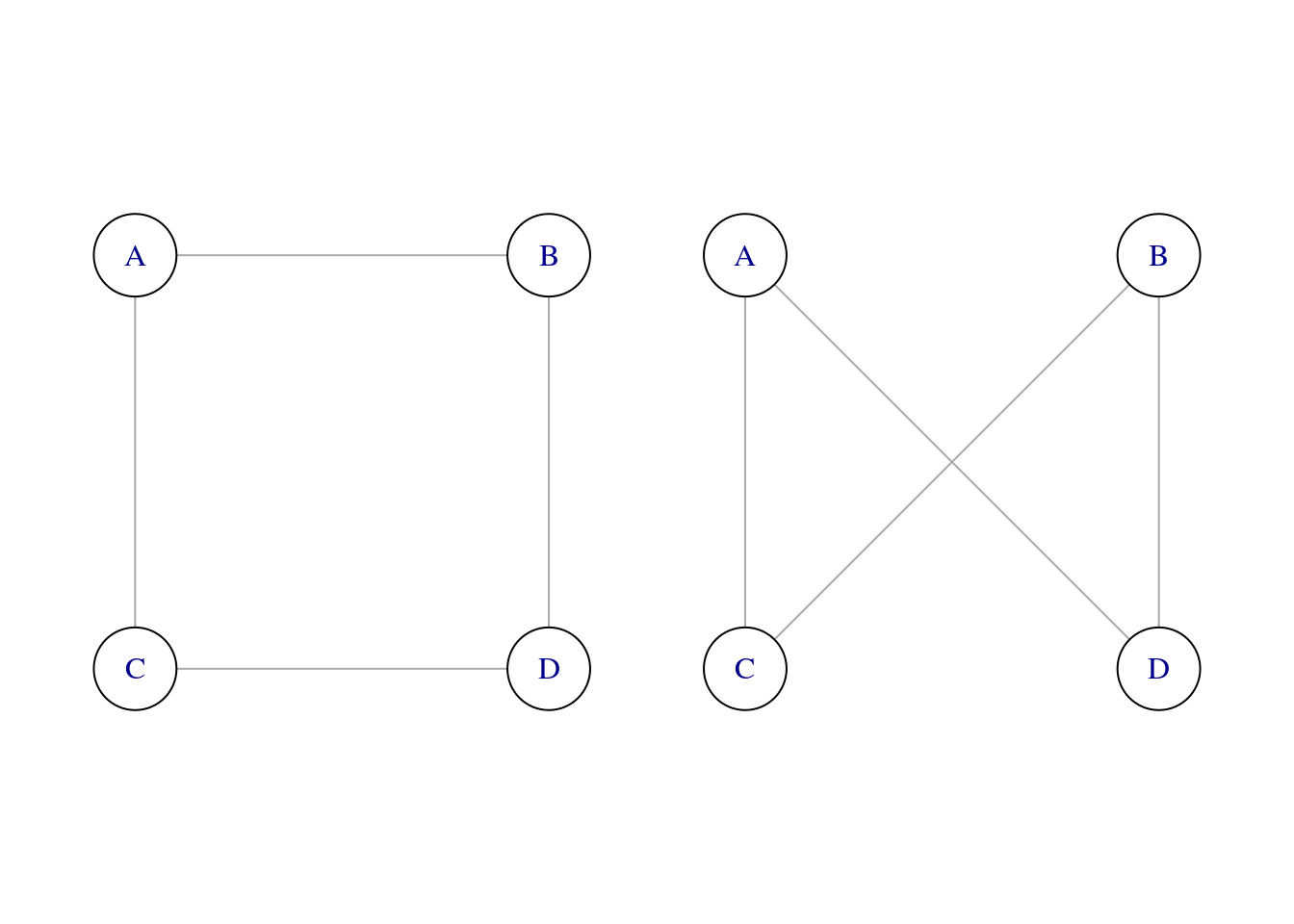
If subgraphs of the same isomorphic classe appear in a network at an abnormal rate, they are be considered a motif of that network. To examine subgraphs, we have several functions in igraph:
motifs(g, n)counts the subgraphs of each isomorphic class of sizen(number of nodes) in a graphg.count_motifs(g, n)returns the total number of subgraphs of sizening.
To illustrate this, let’s consider a power grid network:
power_grid## IGRAPH f3f2e8d UN-- 4941 6594 --
## + attr: id (v/c), name (v/c)
## + edges from f3f2e8d (vertex names):
## [1] 6 --8 7 --8 8 --9 9 --10 5 --13 12--13 13--14 13--15 14--16
## [10] 14--17 18--19 19--20 14--29 33--34 34--35 35--36 36--37 36--38
## [19] 36--39 41--42 36--47 44--47 45--47 46--47 10--50 21--51 24--53
## [28] 25--58 41--58 58--59 59--60 9 --61 31--62 63--68 64--68 65--68
## [37] 35--69 66--69 68--69 67--70 69--70 72--73 9 --75 48--75 76--77
## [46] 78--79 79--80 81--82 13--83 71--86 76--86 4 --88 55--88 87--88
## [55] 87--89 88--90 73--91 11--94 26--94 43--94 92--94 93--94 94--95
## [64] 34--97 40--97 28--98 42--98 57--98 74--98 82--98 98--100 99--100
## + ... omitted several edgesLet’s apply the two functions to power_grid for subgraphs of size n = 3 nodes.
motifs(power_grid, 3) # four motifs of size 3## [1] NA NA 16980 651count_motifs(power_grid, 3) # total number of motifs of a given size in igraph ## [1] 17631As seen in motifs() each isomorphism class has a number, corresponding to its classification in igraph. To find a representative of a isomorphism class, we have the function graph_from_isomorphism_class(). From the result of motifs() we have learned that there are four isomorphism classes of size three. These are labelled from zero to three. Let’s obtain each of the representatives:
graph3_list <- map(0:3, ~ graph_from_isomorphism_class(3, ., directed = FALSE))
titles3 <- paste0("isomorphism ", 0:3)Let’s define a layout for a graph of three nodes.
layout3 <- matrix(c(0, 2,
1, 1,
0, 0), nrow = 3, byrow = TRUE)And here are each of the representatives of each of the isomorphic classes of size three.
par(mfrow = c(2, 2), oma = c(0.5, 1, 0.5, 1), mar = c(1, 1, 1, 1))
walk2(graph3_list, titles3, ~ plot(.x, vertex.size = 40, vertex.color = "white", layout = layout3, main = .y))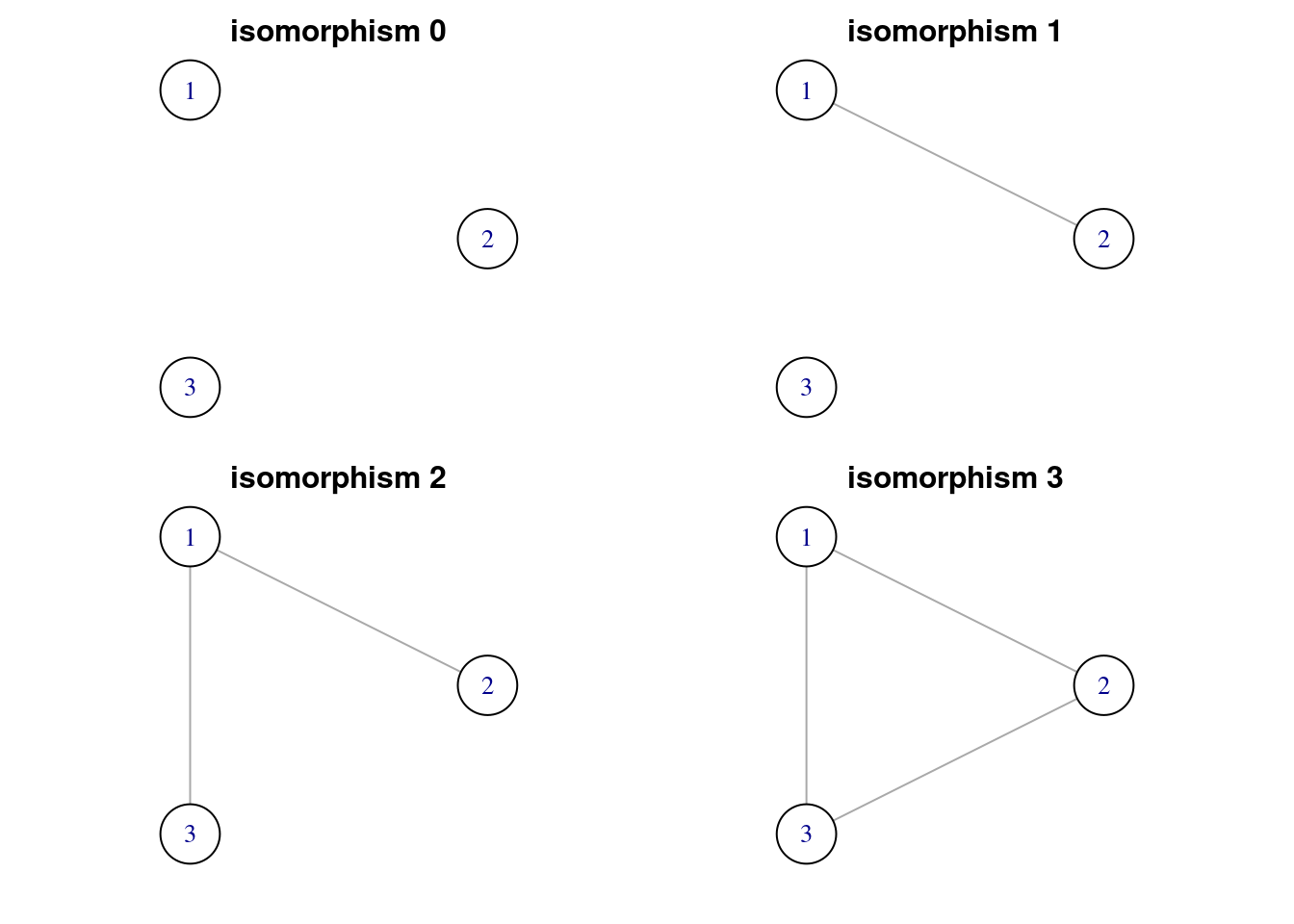
When we have applied the motifs() function to the graph, some of the results are NA. These correspond to unconnected graphs that are not considered as candidates to motifs of order three. Then the possible motifs of order three are:
motifs3_list <- graph3_list[which(!is.na(motifs(power_grid, 3)))]
titles_motifs3 <- titles3[which(!is.na(motifs(power_grid, 3)))]
par(mfrow = c(1, 2), oma = c(0.5, 1, 0.5, 1), mar = c(1, 1, 1, 1))
walk2(motifs3_list, titles_motifs3, ~ plot(.x, vertex.size = 40, vertex.color = "white", layout = layout3, main = .y))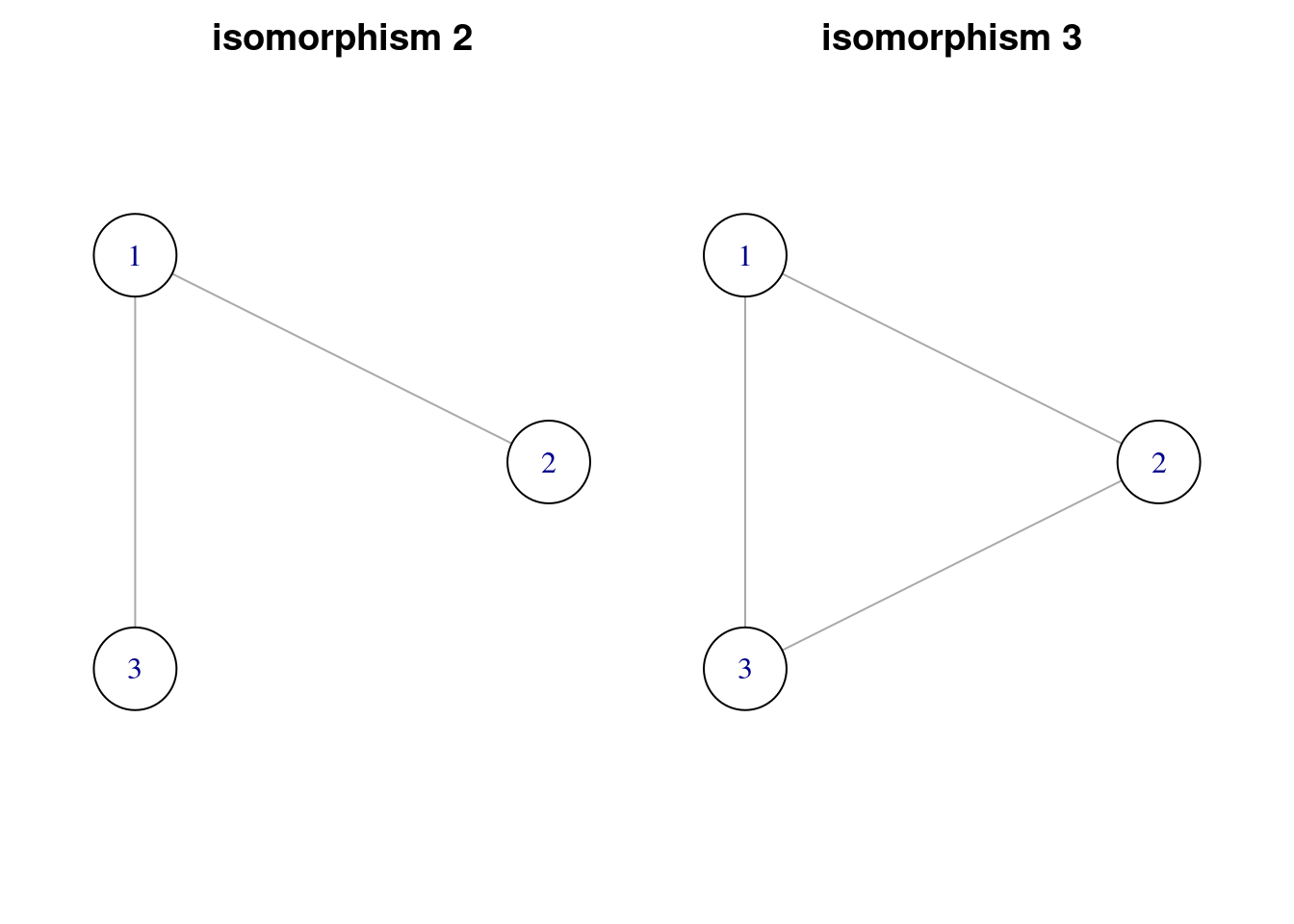
We can repeat the same workflow to obtain the eleven isomorphic classes of size four. Let’s start counting motifs of the sample graph.
motifs(power_grid, 4)## [1] NA NA NA NA 19826 NA 37682 5094 324 385 90There are eleven isomorphic classes:
graph4_list <- map(0:10, ~ graph_from_isomorphism_class(4, ., directed = FALSE))
numbers4 <- substr(as.character(100:110), 2, 3)
titles4 <- paste0("isomorphism ", numbers4)
layout4 <- matrix(c(0, 1,
1, 1,
0, 0,
1, 0), nrow = 4, byrow = TRUE)
par(mfrow = c(4, 3), oma = c(0.5, 1, 0.5, 1), mar = c(1, 1, 1, 1))
walk2(graph4_list, titles4, ~ plot(.x, vertex.size = 40, vertex.color = "white", layout = layout4, main = .y))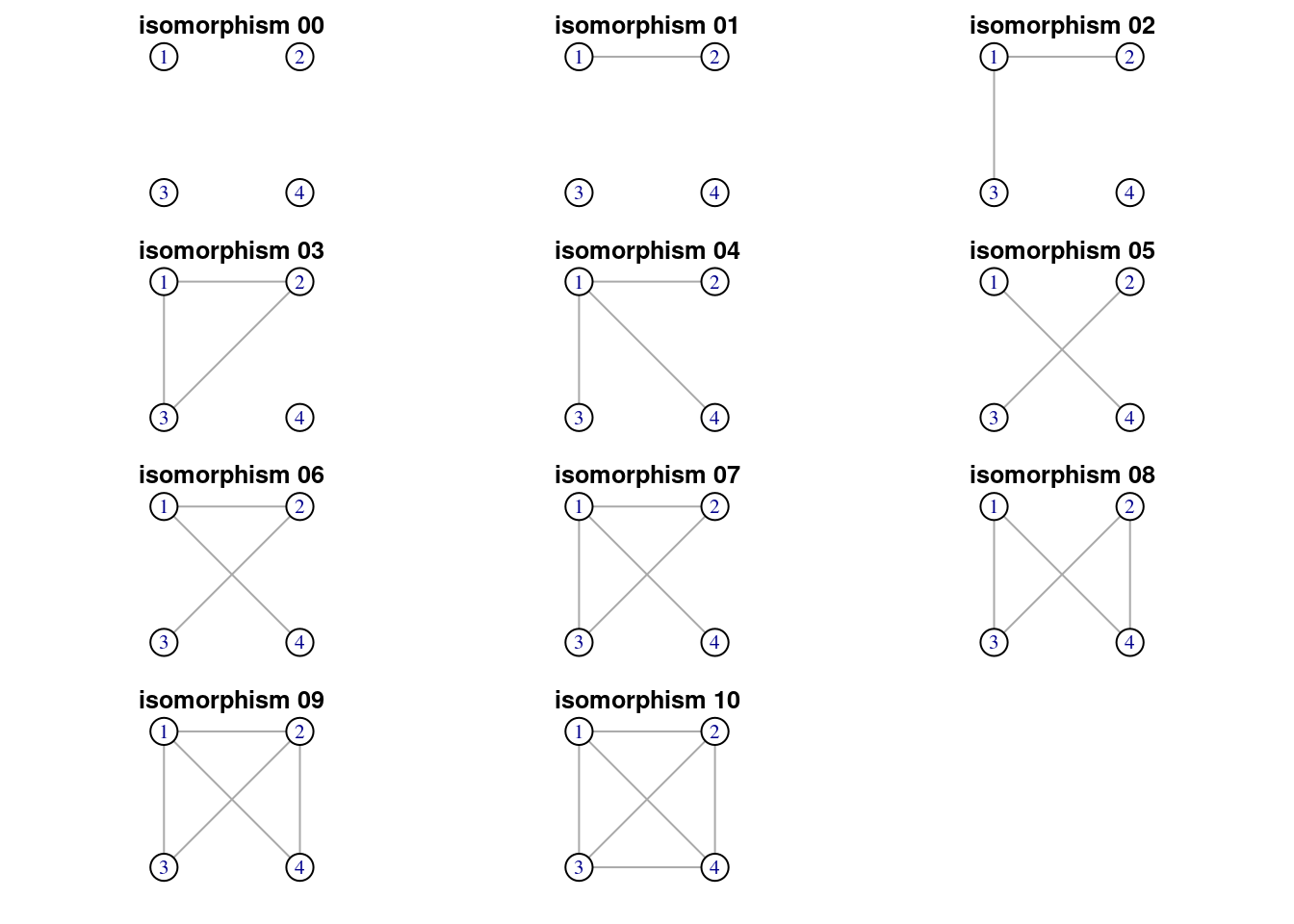
Of the eleven isomorphic classes, only six correspond to possible network motifs.
motifs4_list <- graph4_list[which(!is.na(motifs(power_grid, 4)))]
titles_motifs4 <- titles4[which(!is.na(motifs(power_grid, 4)))]
par(mfrow = c(2, 3), oma = c(0.5, 1, 0.5, 1), mar = c(1, 1, 1, 1))
walk2(motifs4_list, titles_motifs4, ~ plot(.x, vertex.size = 40, vertex.color = "white", layout = layout4, main = .y))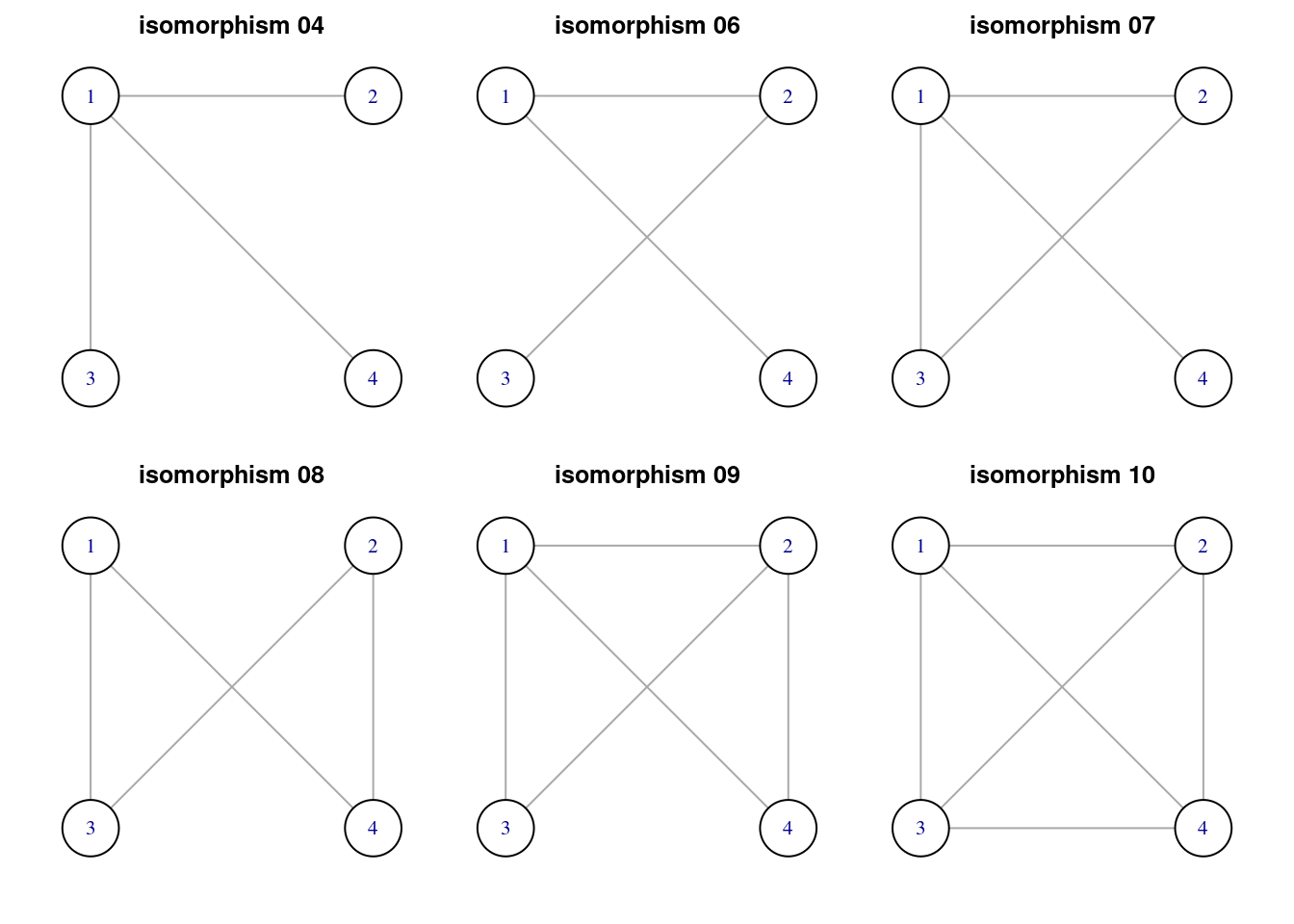
Let’s count the isomorphic classes of size five.
motifs(power_grid, 5)## [1] NA NA NA NA NA NA NA NA NA NA
## [11] NA 25101 NA 118571 8616 82780 12036 3171 1926 NA
## [21] 11703 1785 785 23 107 818 311 355 315 30
## [31] 215 8 23 15There are 34 isomorphic classes of size five, of which 21 are considered connected and then candidates to motifs. Let’s plot the possible motifs of size five.
graph5_list <- map(0:33, ~ graph_from_isomorphism_class(5, ., directed = FALSE))
numbers5 <- substr(as.character(100:133), 2, 3)
titles5 <- paste0("isomorphism ", numbers5)
m5 <- which(!is.na(motifs(power_grid, 5)))
motifs5_list <- graph5_list[m5]
titles_motifs5 <- titles5[m5]
layout5 <- matrix(c(cos(2*pi*0:4/5 + pi/2),
sin(2*pi*0:4/5 + pi/2)), nrow = 5)
par(mfrow = c(7, 3), oma = c(0.5, 1, 0.5, 1), mar = c(1, 1, 1, 1))
walk2(motifs5_list, titles_motifs5, ~ {plot(.x,
vertex.size = 40, vertex.color = "white",
layout = layout5)
title(.y, cex.main = 0.8)})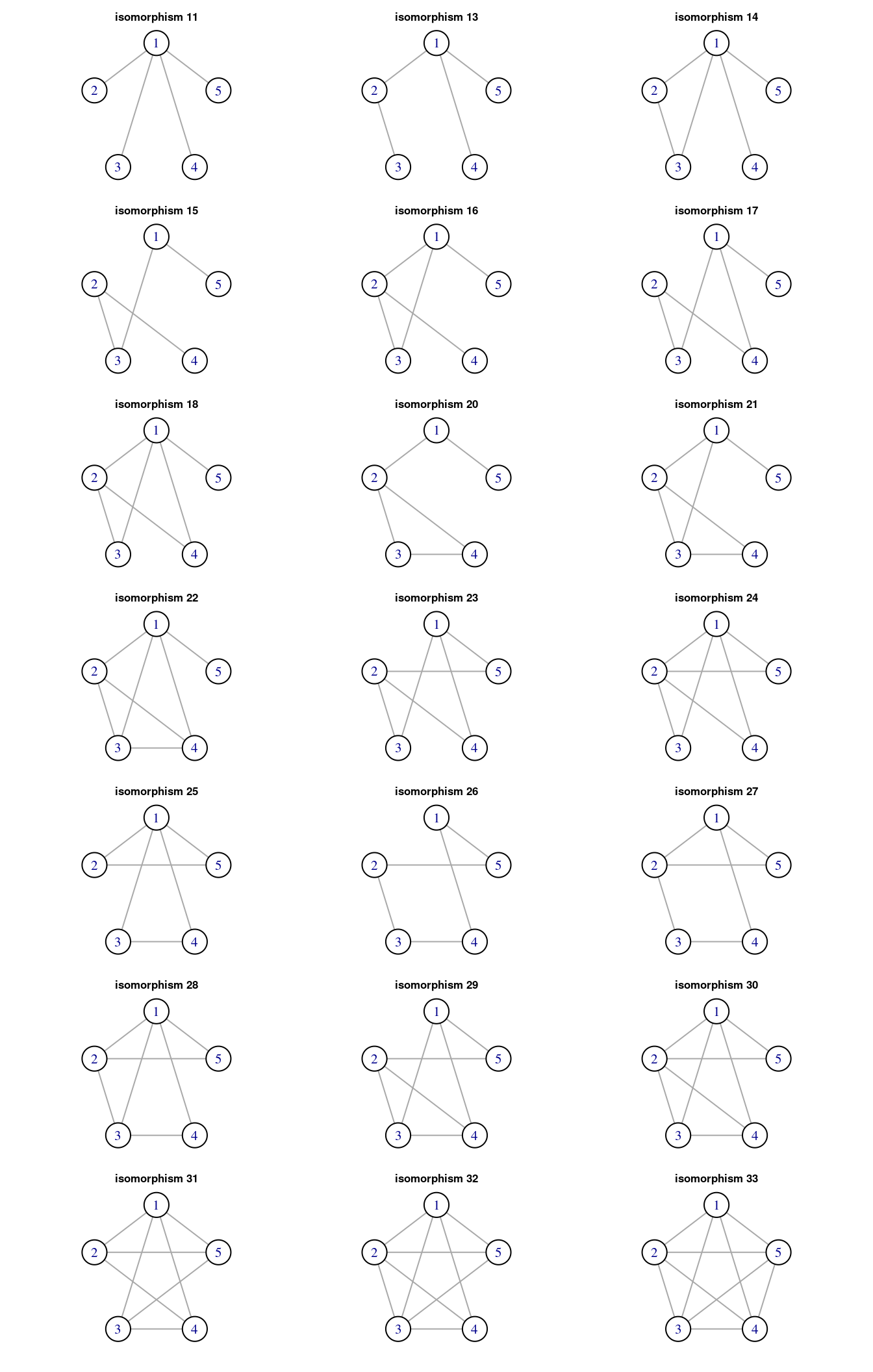
For undirected graphs, igraph goes as far as subgraphs of size six:
motifs(power_grid, 6)## [1] NA NA NA NA NA NA NA NA NA NA
## [11] NA NA NA NA NA NA NA NA NA NA
## [21] NA NA NA NA NA NA NA NA NA NA
## [31] NA NA NA NA 33749 NA 179144 12571 NA 86407
## [41] 257075 38765 5498 3077 NA 19350 2541 983 4440 2224
## [51] NA NA 241372 8020 NA 35826 180917 6734 53371 7820
## [61] 2078 2225 132 416 2450 3530 1592 698 26523 7312
## [71] 1601 1411 125 814 NA 6964 138 NA 4384 630
## [81] 738 124 4273 1448 879 19 72 63 128 NA
## [91] 2053 601 115 132 1 35 558 571 63 73
## [101] 24 170 0 59 331 356 175 27 76 275
## [111] 88 14 49 152 4 22 1 18 0 30
## [121] NA 2000 826 177 188 73 299 27 61 137
## [131] 180 25 11 49 48 0 49 0 1 0
## [141] 0 2 26 0 6 9 10 0 12 2
## [151] 1 13 0 0 0 2We have up to 156 subgraphs of size six, of which which(!is.na(motifs(power_grid, 6))) |> length() are considered motifs.
Table with Isomorphic Classes
To count all isomorphic classes of subgraphs available in igraph, I have defined the motif_count() function. It takes a graph g as argument and returns a data table with all subgraphs of size n belonging to isomorphic class m.
motif_count <- function(g){
motif_count <- map_dfr(3:6, ~ {
mc <- motifs(g, .)
df <- data.table(n = ., m = 0:(length(mc) - 1), mc = mc)
df <- df[!is.na(mc)]
})
return(motif_count)
}This is the result of applying the function to the power grid:
motif_count(power_grid)## n m mc
## <int> <int> <num>
## 1: 3 2 16980
## 2: 3 3 651
## 3: 4 4 19826
## 4: 4 6 37682
## 5: 4 7 5094
## ---
## 137: 6 151 13
## 138: 6 152 0
## 139: 6 153 0
## 140: 6 154 0
## 141: 6 155 2Detecting Motifs in a Graph
To detect the motifs of the power grid, we need to compare the frequency of each subgraph with the one of random graphs with the same number of nodes, edges and degree sequence. The later is obtained with degree().
degree_pg <- degree(power_grid)We can generate random graphs with a fixed degree sequence with the sample_degseq() function. I am using using the Viger and Latapy method passing method = "vl".
set.seed(55)
sample_degseq(degree_pg, method = "vl")## IGRAPH f6b8f59 U--- 4941 6594 -- Degree sequence random graph
## + attr: name (g/c), method (g/c)
## + edges from f6b8f59:
## [1] 1--1017 1--1502 1--2461 2--3785 2--2982 2-- 10 2--2822 3--2881
## [9] 4-- 790 5-- 545 6--4405 6--1624 7--1105 8--3926 9--3227 9--3873
## [17] 9--2198 10--3956 10-- 638 10--1311 10-- 338 10-- 795 11--3839 11-- 670
## [25] 12-- 637 12-- 216 13--1334 14--4861 14--3828 14--3774 14--2844 14--3187
## [33] 15--4591 15--1270 15--3085 15--4148 15--1981 16--4479 17--1050 18--4436
## [41] 19--2315 19--2329 20-- 142 20--2675 20--1952 21--2393 21--1194 22--4873
## [49] 22--3526 23--2535 24--1290 25--2940 25--1045 26--1392 26--3602 27-- 817
## [57] 27--4795 27--2802 27--3838 28--1202 28--1469 29-- 131 30--2503 30--1879
## + ... omitted several edgesTo detect network motifs, I am using the method described in Milo et al. (2002) and Milo et al. (2004). From a sample of random graphs with the same degree sequence we obtain the average and standard deviation of the number of appearances of each motif . Then, I compute the z-score for motif :
Where and are the mean and the standard deviation of number of motifs , respectively. As is normally distributed, network motifs will have values of above the upper significance threshold. We can also speak of antimotifs, with values of below than the lower significance threshold. Antimotifs are subgraphs that appear with a lesser frequence than in an equivalent random network.
To perform this test I have written the motif_detector() function. In addition to the graph g, takes as arguments:
sample_size: the number of random graphs to be evaluated.sig_level: the level of significance to detect motifs and antimotifs.
motif_detector <- function(g, sample_size, sig_level = 0.01){
degseq <- degree(g)
# sampling motifs
random_sg <- map_dfr(1:sample_size, ~{
# random sample graph
rs <- sample_degseq(degseq, method = "vl")
mc <- motif_count(rs)
})
rt <- random_sg[, .(mean = mean(mc), sd = sd(mc)), .(n, m)]
# motifs of g
rn <- motif_count(g)
setnames(rn, "mc", "mc_real")
# merging and computing z score
md <- merge(rt, rn, by = c("n", "m"))
md[, z := (mc_real - mean)/sd]
# significance thresholds
lt <- qnorm(sig_level)
ut <- qnorm(1 - sig_level)
md[, motif := "no motif"]
md[z < lt, motif := "antimotif"]
md[z > ut, motif := "motif"]
return(md)
}Motifs of the Power Grid Network
Let’s apply this function to the power grid network:
mpg <- motif_detector(power_grid, sample_size = 1000)Let’s examine motifs of size three:
mpg[n == 3]## Clave <n, m>
## n m mean sd mc_real z motif
## <int> <int> <num> <num> <num> <num> <char>
## 1: 3 2 18921.849 5.851704 16980 -331.8433 antimotif
## 2: 3 3 3.717 1.950568 651 331.8433 motifThe closed triangle (n = 3, m = 3) is a motif of this network.
mpg[n == 4]## Clave <n, m>
## n m mean sd mc_real z motif
## <int> <int> <num> <num> <num> <num> <char>
## 1: 4 4 26003.758 26.440774 19826 -233.64512 antimotif
## 2: 4 6 53233.384 522.243674 37682 -29.77802 antimotif
## 3: 4 7 46.204 26.394792 5094 191.24212 motif
## 4: 4 8 7.807 2.829798 324 111.73694 motif
## 5: 4 9 0.019 0.136593 385 2818.45261 motif
## 6: 4 10 0.000 0.000000 90 Inf motifHigher-order isomorphic classes are motifs of this network. This suggests that this network has more clustered regions than a random network of similar degree distribution.
References
- Graph motifs (igraph R manual): https://igraph.org/r/doc/motifs.html
- Graph isomorphism: https://www2.math.upenn.edu/~mlazar/math170/notes05-2.pdf
- Milo, R., Itzkovitz, S., Kashtan, N., Levitt, R., Shen-Orr, S., Ayzenshtat, I., … & Alon, U. (2004). Superfamilies of evolved and designed networks. Science, 303(5663), 1538-1542.
- Milo, R., Shen-Orr, S., Itzkovitz, S., Kashtan, N., Chklovskii, D., & Alon, U. (2002). Network motifs: simple building blocks of complex networks. Science, 298(5594), 824-827.
Session Info
## R version 4.5.0 (2025-04-11)
## Platform: x86_64-pc-linux-gnu
## Running under: Linux Mint 21.1
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.10.0
## LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.10.0 LAPACK version 3.10.0
##
## locale:
## [1] LC_CTYPE=es_ES.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=es_ES.UTF-8 LC_COLLATE=es_ES.UTF-8
## [5] LC_MONETARY=es_ES.UTF-8 LC_MESSAGES=es_ES.UTF-8
## [7] LC_PAPER=es_ES.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=es_ES.UTF-8 LC_IDENTIFICATION=C
##
## time zone: Europe/Madrid
## tzcode source: system (glibc)
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] data.table_1.17.0 purrr_1.0.4 igraph_2.1.4
##
## loaded via a namespace (and not attached):
## [1] vctrs_0.6.5 cli_3.6.4 knitr_1.50 rlang_1.1.6
## [5] xfun_0.52 generics_0.1.3 jsonlite_2.0.0 glue_1.8.0
## [9] htmltools_0.5.8.1 sass_0.4.10 rmarkdown_2.29 tibble_3.2.1
## [13] evaluate_1.0.3 jquerylib_0.1.4 fastmap_1.2.0 yaml_2.3.10
## [17] lifecycle_1.0.4 bookdown_0.43 compiler_4.5.0 dplyr_1.1.4
## [21] pkgconfig_2.0.3 rstudioapi_0.17.1 blogdown_1.21 digest_0.6.37
## [25] R6_2.6.1 tidyselect_1.2.1 pillar_1.10.2 magrittr_2.0.3
## [29] bslib_0.9.0 tools_4.5.0 cachem_1.1.0