In frequentist statistics, a confidence interval is an interval which is expected to typically contain the population parameter being estimated. More specifically, given a confidence level , a confidence interval is a random interval that contains the population parameter with probability . The typical values of the confidence level of the interval are 0.95 and 0.99. The confidence interval is an alternative metric to -value to obtain information about a population parameter trough hypothesis testing.
In this post, I will illustrate how confidence intervals work in the estimation of the mean of a random variable. I will use the tidyverse and the broom package to obtain the output of statistical test in tidy format:
library(tidyverse)
library(broom)Calculating Confidence Intervals
Let’s start obtainining a sample of a normal distribution with mean zero and standard deviation one with the rnorm() function. If I want a sample of 100 observations we proceed as follows:
set.seed(1212)
x <- rnorm(100)Let’s use t.test() to check if the population mean is zero:
t.test(x)##
## One Sample t-test
##
## data: x
## t = -0.84443, df = 99, p-value = 0.4005
## alternative hypothesis: true mean is not equal to 0
## 95 percent confidence interval:
## -0.2835738 0.1142651
## sample estimates:
## mean of x
## -0.08465435We observe that:
- The p-value of the hypothesis testing is higher than the standard threshold of 0.05.
- The confidence interval contains the hypothesized population mean of zero.
Therefore, we cannot reject the null hypothesis that the mean is equal to zero.
What would happen if we perform this tests to many samples from the same distribution? If we want to store them in a data frame, we might want to use broom::tidy() to obtain the result in a tibble format:
tidy(t.test(x))## # A tibble: 1 × 8
## estimate statistic p.value parameter conf.low conf.high method alternative
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
## 1 -0.0847 -0.844 0.400 99 -0.284 0.114 One Sampl… two.sidedLet’s wrap it together in a conf_int() function that takes the sample size n as input and:
- generates a sample from a normal distribution with
rnorm(). - performs the
t.test()of the sample mean. - stores the output in tidy format, retaining the variables of interest: the mean
estimate, the lower and upper bound of the confidence intervalconf.lowandconf.highand thep.value.
conf_int <- function(n = 100, mean = 0, sd = 1){
sample <- rnorm(n = n, mean = mean, sd = sd)
test <- t.test(sample)
result <- tidy(test) |>
select(estimate, conf.low, conf.high, p.value)
return(result)
}A run of the function leads to:
set.seed(1212)
conf_int()## # A tibble: 1 × 4
## estimate conf.low conf.high p.value
## <dbl> <dbl> <dbl> <dbl>
## 1 -0.0847 -0.284 0.114 0.400Now we can use purrr::map_dfr() to wrap the results of many runs of conf_int() in a single tibble. Once obtained, we can check whether zero is within the confidence interval and store the result in a column, and give and id to each observation. All of this is done in the set_intervals() function, that takes as arguments n and the number of samples sample.
set_intervals <- function(sample = 100, n = 100, mean = 0, sd = 1){
intervals <- map_dfr(1:sample, ~ conf_int(n = n, mean = mean, sd = sd))
intervals <- intervals |>
mutate(id = 1:n(),
result = ifelse(sign(conf.low) == sign(conf.high), "accept", "reject")) |>
relocate(id)
return(intervals)
}The outcome of the function looks like:
set.seed(1111)
intervals <- set_intervals()
intervals## # A tibble: 100 × 6
## id estimate conf.low conf.high p.value result
## <int> <dbl> <dbl> <dbl> <dbl> <chr>
## 1 1 0.242 0.0225 0.461 0.0310 accept
## 2 2 0.210 -0.00567 0.425 0.0562 reject
## 3 3 -0.0705 -0.250 0.109 0.438 reject
## 4 4 -0.0869 -0.269 0.0952 0.346 reject
## 5 5 -0.0155 -0.215 0.184 0.878 reject
## 6 6 -0.190 -0.373 -0.00657 0.0425 accept
## 7 7 -0.0926 -0.290 0.105 0.354 reject
## 8 8 -0.281 -0.478 -0.0847 0.00549 accept
## 9 9 -0.0622 -0.262 0.138 0.539 reject
## 10 10 0.0850 -0.0901 0.260 0.338 reject
## # ℹ 90 more rowsPlotting many Confidence Intervals
Let’s plot the results, using geom_point() for the sample mean and geom_segment():
intervals |>
ggplot(aes(estimate, id, color = result)) +
geom_point() +
geom_segment(aes(x = conf.low, y = id, xend = conf.high, yend = id, color = result))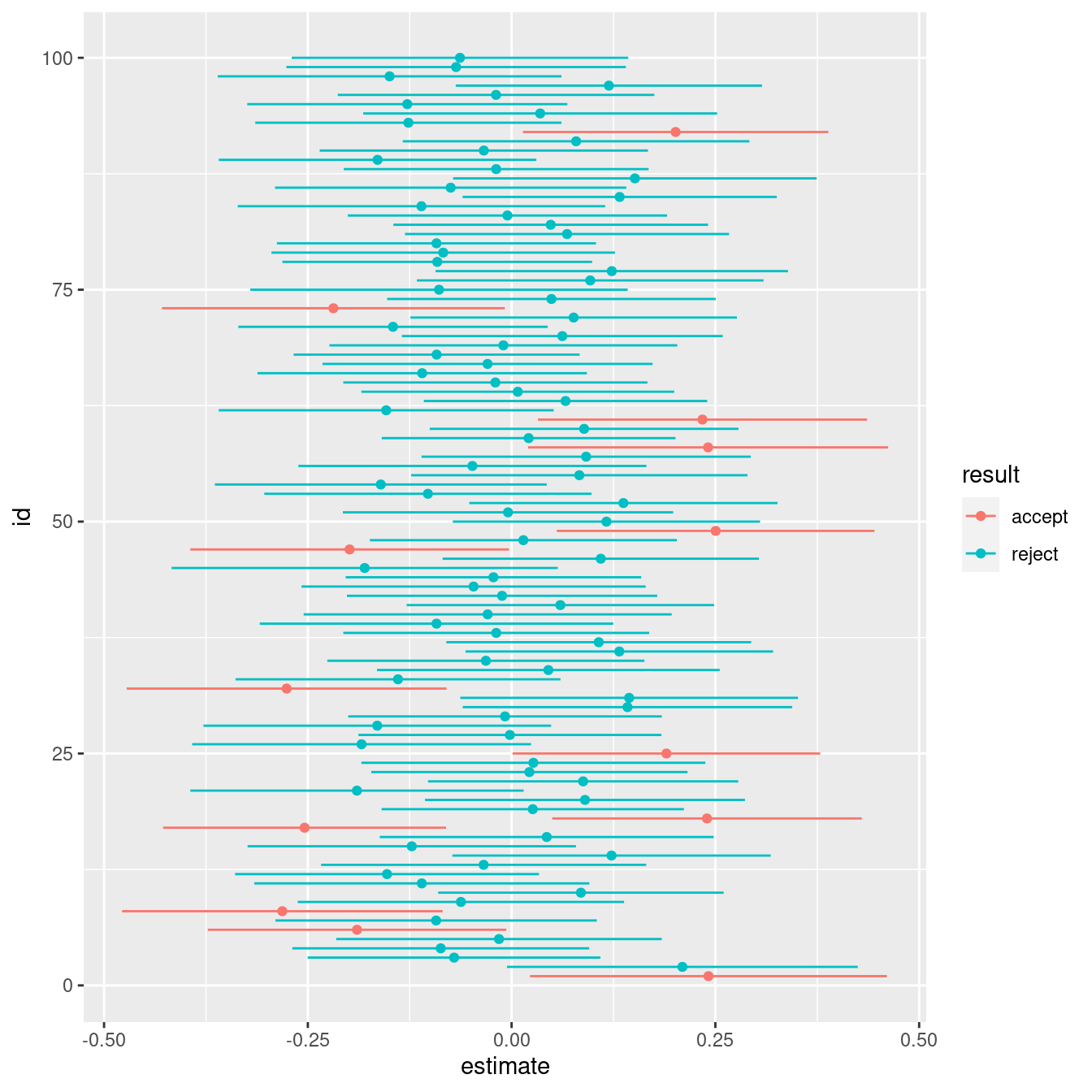
A nicer version of the same plot:
intervals |>
ggplot(aes(estimate, id, color = result)) +
geom_point() +
geom_segment(aes(x = conf.low, y = id, xend = conf.high, yend = id, color = result)) +
geom_vline(xintercept = 0, linetype = "dashed") +
ggtitle("Several confidence intervals") +
scale_color_manual(values = c("#FF3333", "#009900")) +
theme_minimal() +
theme(axis.text.y = element_blank(),
axis.title.y = element_blank(),
axis.title.x = element_blank(),
legend.position = "none",
plot.title.position = "plot")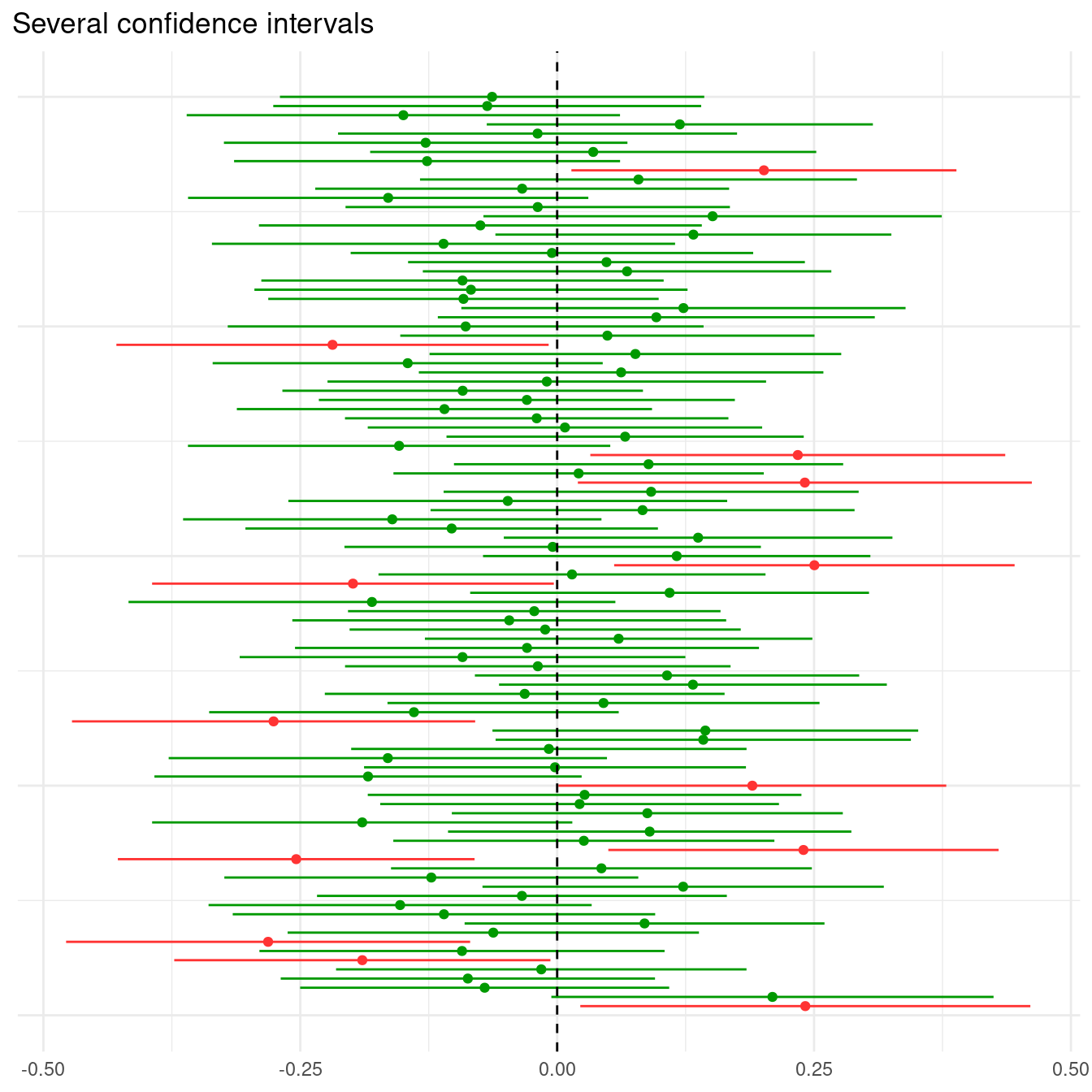
In this case, the population that we are taking the samples from has a mean equal to zero, so that the null hypothesis that the population mean is equal is zero is true. Therefore, we observe that most of the confidence intervals contain the zero (in green), but some of them do not (in red).
intervals |>
group_by(result) |>
count()## # A tibble: 2 × 2
## # Groups: result [2]
## result n
## <chr> <int>
## 1 accept 13
## 2 reject 87Specifically, in 13 out of the 100 intervals, we have rejected the null hypothesis, comiting a Type I error. Let’s relate this with p-values evaluating the minimum and maximum values for the two values of result:
intervals |>
group_by(result) |>
summarise(min_pvalue = min(p.value), max_pvalue = max(p.value))## # A tibble: 2 × 3
## result min_pvalue max_pvalue
## <chr> <dbl> <dbl>
## 1 accept 0.00456 0.0486
## 2 reject 0.0562 0.982In all cases that the confidence interval does not include zero, the p-value is smaller than 0.05. We observe, though, that this is happening 13% of the time, more than the expected 5%. Let’s pick a set of 1,000 samples to get a more precise estimation of probability.
set.seed(3333)
intervals_large <- set_intervals(sample = 1000)Let’s see in how many samples of intervals_large we are committing a Type I error:
intervals_large |>
group_by(result) |>
count()## # A tibble: 2 × 2
## # Groups: result [2]
## result n
## <chr> <int>
## 1 accept 48
## 2 reject 952Now the number of Type I errors is closer to 5%.
Confidence Intervals and Variance
The variability on the original sample affects the confidence interval: the smaller the variance of the same, the smaller the width of the interval. To illustrate this, let’s build a set of intervals taken from a population of smaller variance and compare the results with the previous plot.
set.seed(5555)
intervals_sv <- set_intervals(sd = 0.5)two_intervals <- bind_rows(intervals |> mutate(sd = 1),
intervals_sv |> mutate(sd = 0.5))two_intervals |>
ggplot(aes(estimate, id, color = result)) +
geom_point() +
geom_segment(aes(x = conf.low, y = id, xend = conf.high, yend = id, color = result)) +
facet_grid(. ~ sd, labeller = label_both) +
geom_vline(xintercept = 0, linetype = "dashed") +
ggtitle("Effect of sd on confidence intervals") +
scale_color_manual(values = c("#FF3333", "#009900")) +
theme_minimal() +
theme(axis.text.y = element_blank(),
axis.text.x = element_text(size = 6, angle = 90),
axis.title.y = element_blank(),
axis.title.x = element_blank(),
legend.position = "none",
plot.title.position = "plot",
strip.text.x = element_text(size = 10),
strip.background.x = element_rect(fill = "#FFB266", color = "#808080"))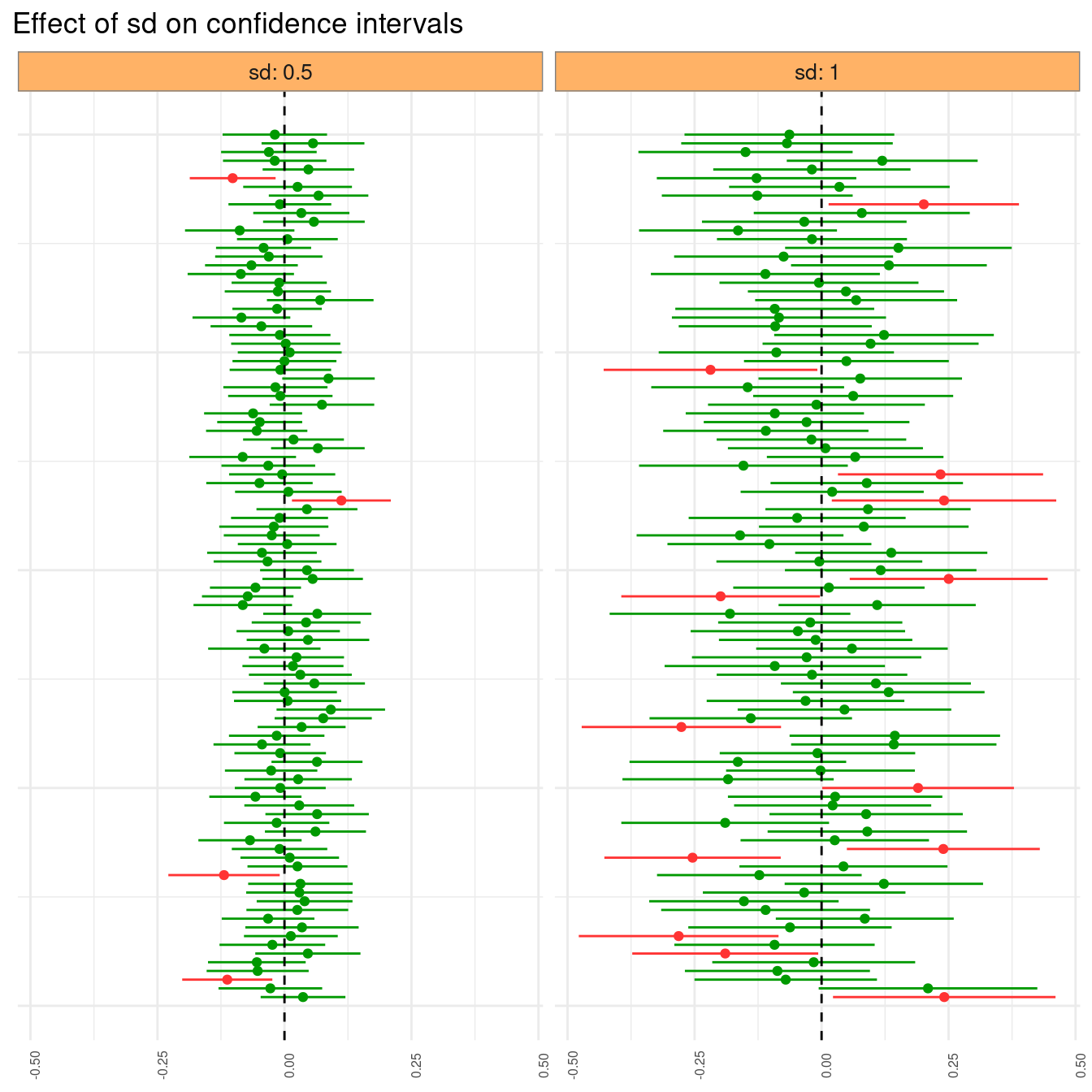
Interpreting Confidence Intervals
In frequentist statistics, a confidence interval is a random interval that contains the (true) population parameter with a probability . The value is the confidence level of the interval.
We can use confidence intervals for hypothesis testing: if the interval does not contain the population parameter defined in the null hypothesis, we can reject the null hypothesis with a probability of Type I error of . The confidence interval is related with the p-value: if the confidence interval does not contain the expected population parameter, the p-value will be smaller than .
Session Info
## R version 4.3.3 (2024-02-29)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Linux Mint 21.1
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.10.0
## LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.10.0
##
## locale:
## [1] LC_CTYPE=es_ES.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=es_ES.UTF-8 LC_COLLATE=es_ES.UTF-8
## [5] LC_MONETARY=es_ES.UTF-8 LC_MESSAGES=es_ES.UTF-8
## [7] LC_PAPER=es_ES.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=es_ES.UTF-8 LC_IDENTIFICATION=C
##
## time zone: Europe/Madrid
## tzcode source: system (glibc)
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] broom_1.0.5 lubridate_1.9.3 forcats_1.0.0 stringr_1.5.1
## [5] dplyr_1.1.4 purrr_1.0.2 readr_2.1.5 tidyr_1.3.0
## [9] tibble_3.2.1 ggplot2_3.4.4 tidyverse_2.0.0
##
## loaded via a namespace (and not attached):
## [1] sass_0.4.5 utf8_1.2.3 generics_0.1.3 blogdown_1.16
## [5] stringi_1.7.12 hms_1.1.3 digest_0.6.31 magrittr_2.0.3
## [9] evaluate_0.20 grid_4.3.3 timechange_0.2.0 bookdown_0.33
## [13] fastmap_1.1.1 jsonlite_1.8.8 backports_1.4.1 fansi_1.0.4
## [17] scales_1.2.1 jquerylib_0.1.4 cli_3.6.1 rlang_1.1.3
## [21] munsell_0.5.0 withr_2.5.0 cachem_1.0.7 yaml_2.3.7
## [25] tools_4.3.3 tzdb_0.3.0 colorspace_2.1-0 vctrs_0.6.4
## [29] R6_2.5.1 lifecycle_1.0.3 pkgconfig_2.0.3 pillar_1.9.0
## [33] bslib_0.6.1 gtable_0.3.3 glue_1.6.2 highr_0.10
## [37] xfun_0.39 tidyselect_1.2.0 rstudioapi_0.15.0 knitr_1.42
## [41] farver_2.1.1 htmltools_0.5.7 rmarkdown_2.21 labeling_0.4.2
## [45] compiler_4.3.3