In mathematics, a random walk is a succession of random steps on some mathematical space. A popular random walk model is that of a random walk on a regular lattice, where at each step the location jumps to another site according to some probability distribution. In a simple random walk, the location can only jump to neighboring sites of the lattice, forming a lattice path.
In this post, I will present a function to generate a two-dimensional random walk and another function to plot it. This exercise will allow me to present an example of vectorial programming and some functionalities of the tidyverse.
library(tidyverse)Generating Lattice Random Walks
Here is a function random_walk() that returns the evolution of a random walk for a number of steps in a lattice of dimension d. The setup values are 100 steps in a 2-dimensional lattice.
random_walk <- function(steps = 100, d = 2){
dim <- sample(1:d, steps, replace = TRUE)
move <- sample(c(-1, 1), steps, replace = TRUE)
moves <- map_dfc(1:d, ~ c(0, cumsum(ifelse(dim == ., move, 0))))
return(moves)
}I have defined the path of the random walk defining randomly the dimension along the lattice of each movement with dim and the orientation along the dimension with move. dim takes values between one and d, and move takes values -1 or 1, representing the two possible opposite directions. Rather than defining those values iteratively, they are defined as vectors of length steps.
moves is a tibble with d columns and steps rows, which is defined as follows: for each of the d dimensions:
- we define a vector with components equal to the component of
moveifdim == d, and zero otherwise. In R, this can be obtained withifelse(dim == d, move, 0). - we use
cumsum()to obtain the position in the dimension at each step doingcumsum(ifelse(dim == d, move, 0)). - we add the position zero at the beginning, as the process starts at the origin of coordinates:
c(0, cumsum(ifelse(dim == d, move, 0))). - we wrap the list of
dvectors into a tibble ofdcolumns usingmap_dfc()frompurrr.
Let’s obtain a random walk of 100 steps in a 2-dimensional lattice:
set.seed(1111)
rw <- random_walk()
rw## # A tibble: 101 × 2
## ...1 ...2
## <dbl> <dbl>
## 1 0 0
## 2 0 -1
## 3 0 0
## 4 0 -1
## 5 0 -2
## 6 0 -3
## 7 -1 -3
## 8 -1 -2
## 9 -1 -3
## 10 -1 -2
## # ℹ 91 more rowsPlotting 2-Dimensional Lattice Random Walks
Let’s define a plot_random_walk() function to plot a two-dimensional random walk.
plot_random_walk <- function(coords){
names(coords) <- c("x0", "y0")
segments <- coords |>
mutate(x1 = lead(x0), y1 = lead(y0)) |>
slice(-n())
segments <- segments |>
group_by(x0, y0, x1, y1) |>
count()
s_segments <- full_join(segments,
segments,
by =c("x0" = "x1", "y0" = "y1",
"x1" = "x0", "y1" = "y0")) |>
replace_na(list(n.x =0, n.y = 0)) |>
mutate(n = n.x + n.y)
ggplot(coords, aes(x0, y0)) +
geom_point() +
geom_segment(data = s_segments, aes(x = x0, y = y0, xend = x1, yend = y1, linewidth = n)) +
scale_linewidth(range = c(1, 3)) +
theme_void() +
theme(legend.position = "none")
}The function takes as input the output of random_walk() and prepares a suitable table ready to be used with ggplot with the following steps:
- Rename the columns of the table as
x0andy0. - Define the
segmentstable with the start (x0andy0) and end (x1andy1) point of each segment. We usedplyr::lead()to obtain the end points of segments andslice(-n())to remove the last row. - Some of the segments may appear more than once along the path. It can be interesting to check how many times appears each segment. We use
dplyr::group_by()anddplyr::count()to redefinesegmentsin this way. The number of occurrences of each segment is stored in the columnngenerated bycount(). - In the table
s_segments, I add the number of occurrences of segments fromx0, y0tox1, y1and of segments fromx1, y1tox0, y0. As I am doing a merge ofsegmentswith itself, the variable n will appear asn.xandn.yin the merged table.
Now we are ready to do the plot:
- with
geom_point()we draw each of the points covered by the random walk usingcoords. - with
geom_segment()we draw each of the steps usigns_segments. Thelinewidthof the step is proportional to the number of times the process crosses it in either direction. That’s why it is included inaes(). scale_linewidth(range = c(1, 3))is used to tune the minimum and maximum line width. This feature is available in recent versions ofggplot2.
plot_random_walk(rw)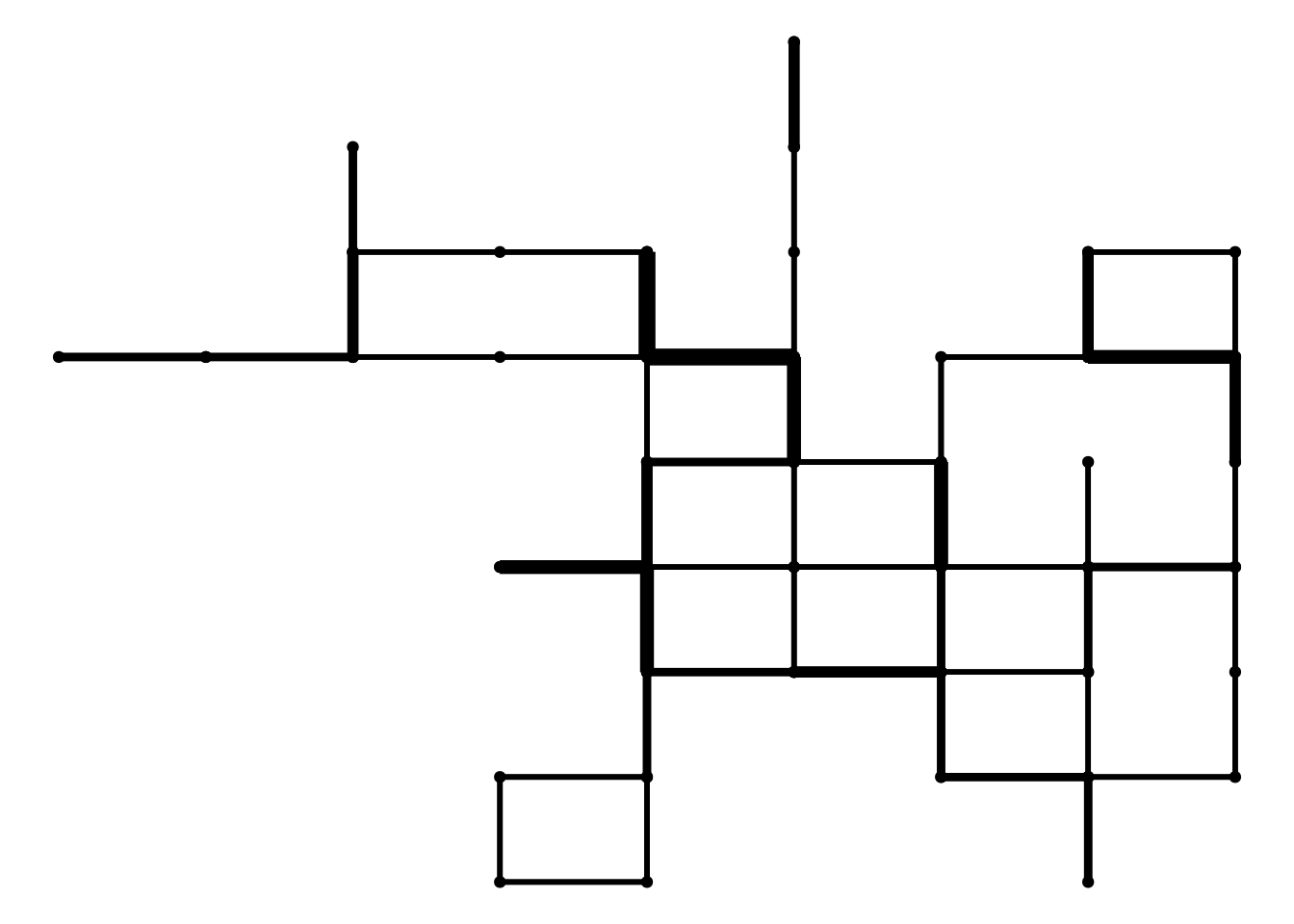
If we set more steps to the random walk, it will cover a much larger area:
set.seed(2222)
rw_1000 <- random_walk(steps = 1000)
plot_random_walk(rw_1000)
A random walk is a succession of random steps on some mathematical space. We can use random walks to model processes like paths of foraging animals, the evolution of a stock price or the financial status of a gambler. In this post, I have used the functionalities of the tidyverse to generate random walks of lattices of arbitrary number of dimensions, and to plot random walk in two-dimensional lattices.
## R version 4.3.2 (2023-10-31)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Linux Mint 21.1
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.10.0
## LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.10.0
##
## locale:
## [1] LC_CTYPE=es_ES.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=es_ES.UTF-8 LC_COLLATE=es_ES.UTF-8
## [5] LC_MONETARY=es_ES.UTF-8 LC_MESSAGES=es_ES.UTF-8
## [7] LC_PAPER=es_ES.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=es_ES.UTF-8 LC_IDENTIFICATION=C
##
## time zone: Europe/Madrid
## tzcode source: system (glibc)
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] lubridate_1.9.2 forcats_1.0.0 stringr_1.5.0 dplyr_1.1.2
## [5] purrr_1.0.1 readr_2.1.4 tidyr_1.3.0 tibble_3.2.1
## [9] ggplot2_3.4.2 tidyverse_2.0.0
##
## loaded via a namespace (and not attached):
## [1] gtable_0.3.3 jsonlite_1.8.4 highr_0.10 compiler_4.3.2
## [5] tidyselect_1.2.0 jquerylib_0.1.4 scales_1.2.1 yaml_2.3.7
## [9] fastmap_1.1.1 R6_2.5.1 labeling_0.4.2 generics_0.1.3
## [13] knitr_1.42 bookdown_0.33 munsell_0.5.0 tzdb_0.3.0
## [17] bslib_0.5.0 pillar_1.9.0 rlang_1.1.0 utf8_1.2.3
## [21] stringi_1.7.12 cachem_1.0.7 xfun_0.39 sass_0.4.5
## [25] timechange_0.2.0 cli_3.6.1 withr_2.5.0 magrittr_2.0.3
## [29] digest_0.6.31 grid_4.3.2 rstudioapi_0.14 hms_1.1.3
## [33] lifecycle_1.0.3 vctrs_0.6.2 evaluate_0.20 glue_1.6.2
## [37] farver_2.1.1 blogdown_1.16 fansi_1.0.4 colorspace_2.1-0
## [41] rmarkdown_2.21 tools_4.3.2 pkgconfig_2.0.3 htmltools_0.5.5