Factor analysis is a statistical technique used to analyze the relationships among a large number of observed variables and to identify a smaller number of underlying, unobserved variables, which are called factors. The goal of factor analysis is to reduce the complexity of a dataset and to identify the underlying structure that explains the observed data.
Factor analysis is used in a variety of fields, including psychology, sociology, economics, marketing, and biology. The results of factor analysis can help researchers to better understand the relationships among the observed variables and to identify the underlying factors that are driving those relationships.
In predictive modelling, factor analysis can be used to reduce the number of variables that are included in a model. The factor scores of observations can then be used as input variables in the predictive model.
By reducing the number of variables in the model, factor analysis can help to overcome problems of overfitting, multicollinearity, and high dimensionality. Overfitting occurs when a model is too complex and fits the training data too closely, which can lead to poor performance on new data. Multicollinearity occurs when two or more variables are highly correlated with each other, which can lead to unstable and unreliable estimates of the model coefficients. High dimensionality occurs when there are too many variables in the model, which can lead to computational inefficiency and reduced model interpretability.
Exploratory factor analysis (EFA) and confirmatory factor analysis (CFA) are two different types of factor analysis that are used for different purposes.
Exploratory factor analysis (EFA) is used to explore and identify the underlying factor structure of a dataset. EFA does not require prior knowledge of the relationships among the observed variables. In EFA, the researcher examines the pattern of correlations among the observed variables to identify the factors that best explain the variation in the data. The researcher does not have any preconceived ideas about the number or structure of the factors that underlie the data, and the number of factors is determined based on statistical criteria. On the other hand, confirmatory factor analysis (CFA) is used to test a specific hypothesis about the underlying factor structure of a dataset. CFA requires prior knowledge of the relationships among the observed variables. In CFA, the researcher specifies a theoretical model of the underlying factor structure, and the model is then tested against the observed data to determine how well it fits the data.
In this post, I will present a workflow to carry out exploratory factor analysis (EFA) in R. I will use the tidyverse for handling and plotting data, kableExtra to present tabular data, corrr for examining correlation matrices, the psych package for functions to carry out EFA and lavaan to retrieve the dataset I will be using in the workflow.
library(tidyverse)
library(corrr)
library(psych)
library(lavaan)
library(kableExtra)The HolzingerSwineford1939 dataset contains 301 observations of mental ability scores. The variables relevant for our analysis are x1 to x9.
hz <- HolzingerSwineford1939 |>
select(x1:x9)
hz |>
slice(1:5) |>
kbl() |>
kable_styling(full_width = FALSE)| x1 | x2 | x3 | x4 | x5 | x6 | x7 | x8 | x9 |
|---|---|---|---|---|---|---|---|---|
| 3.333333 | 7.75 | 0.375 | 2.333333 | 5.75 | 1.2857143 | 3.391304 | 5.75 | 6.361111 |
| 5.333333 | 5.25 | 2.125 | 1.666667 | 3.00 | 1.2857143 | 3.782609 | 6.25 | 7.916667 |
| 4.500000 | 5.25 | 1.875 | 1.000000 | 1.75 | 0.4285714 | 3.260870 | 3.90 | 4.416667 |
| 5.333333 | 7.75 | 3.000 | 2.666667 | 4.50 | 2.4285714 | 3.000000 | 5.30 | 4.861111 |
| 4.833333 | 4.75 | 0.875 | 2.666667 | 4.00 | 2.5714286 | 3.695652 | 6.30 | 5.916667 |
Exploring the Correlation mMatrix
The input of factor analysis is the correlation matrix, and for some indices the number of observations. We can explore correlations with the corrr package:
cor_tb <- correlate(hz)corrr::rearrange groups highly correlated variables closer together, and corrr::rplot visualizes the result (I have used a custom color scale to highlight the results):
cor_tb |>
rearrange() |>
rplot(colors = c("red", "white", "blue"))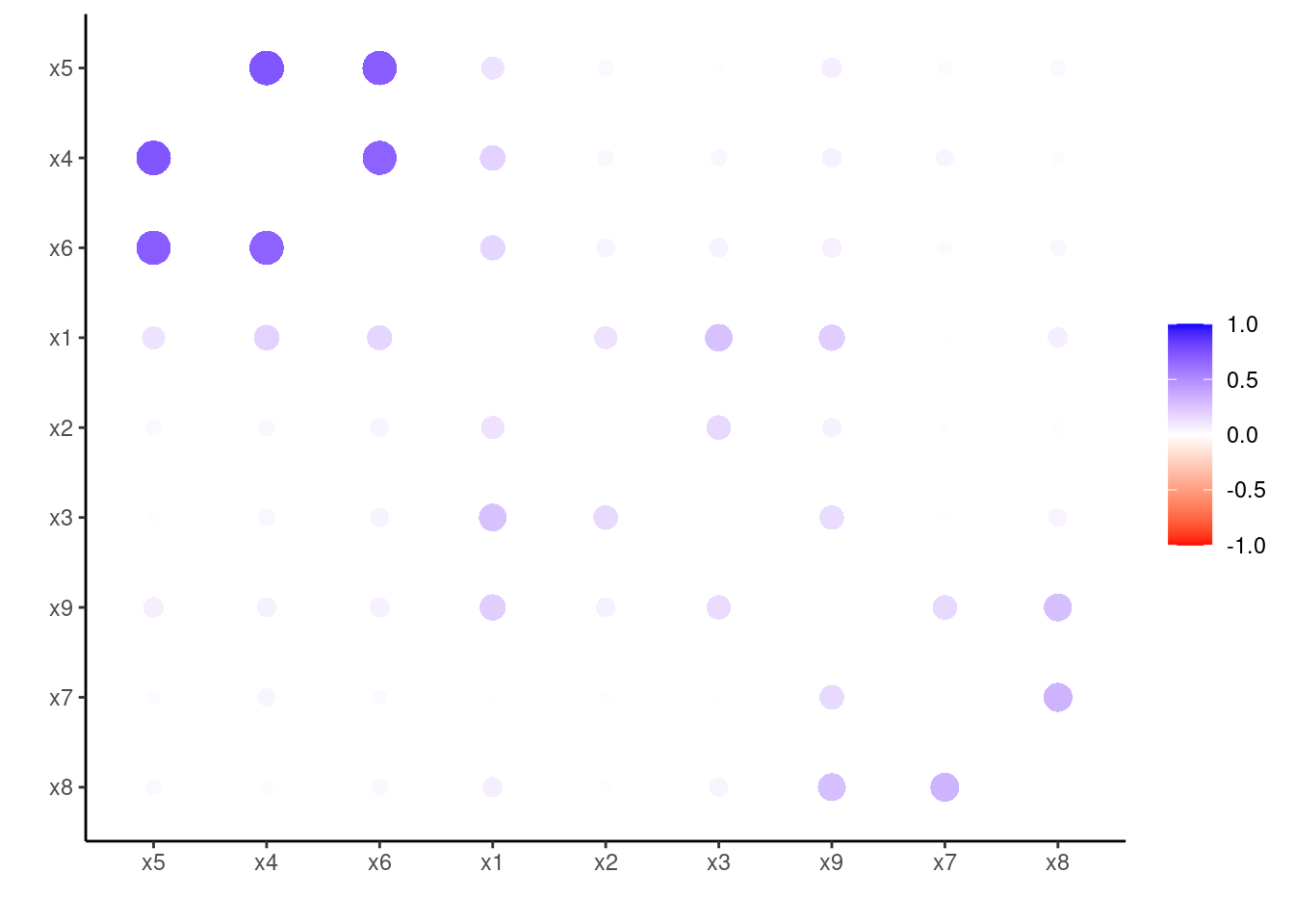
Looks like three groups emerge, around variables:
x4tox6(first group).x1tox3(second group).x7tox9(third group).
Let’s start the factor analysis workflow. To do so, we need to obtain the correlations in matrix format with the R base cor function.
cor_mat <- cor(hz)Preliminary tests
In preliminary analysis, we examine if the correlation matrix is suitable for factor analysis. We have two methods available:
- The Kaiser–Meyer–Olkin (KMO) test of sample adequacy. According to Kaiser (1974), values of KMO > 0.9 are marvelous, in the 0.80s, mertitourious, in the 0.70s, middling, in the 0.60s, mediocre, in the 0.50s, miserable, and less than 0.5, unacceptable. We can obtain that index with
psych::KMO. It also provides a sample adequacy measure for each variable. - The Bartlett’s test of sphericity. It is a testing of the hypothesis that the correlation matrix is an identity matrix. If this null hypothesis can be rejected, we can assume that variables are correlated and then perform factor analysis. We can do this testing with the
psych::cortest.bartlettfunction.
The results of the KMO test are:
KMO(cor_mat)## Kaiser-Meyer-Olkin factor adequacy
## Call: KMO(r = cor_mat)
## Overall MSA = 0.75
## MSA for each item =
## x1 x2 x3 x4 x5 x6 x7 x8 x9
## 0.81 0.78 0.73 0.76 0.74 0.81 0.59 0.68 0.79These eresults can be considered middling. The Bartlett’s test results are:
cortest.bartlett(R = cor_mat, n = 301)## $chisq
## [1] 904.0971
##
## $p.value
## [1] 1.912079e-166
##
## $df
## [1] 36Those results provide strong evidence that variables are correlated.
Number of Factors to Extract
A starting point to decide how many factors to extract is to examine the eigenvalues of correlation matrix.
eigen(cor_mat)$values## [1] 3.2163442 1.6387132 1.3651593 0.6989185 0.5843475 0.4996872 0.4731021
## [8] 0.2860024 0.2377257There are two criteria used to decide the number of factors to extract:
- Pick as many factors as eigenvalues greater than one.
- Examine the elbow point of the scree plot. In that plot we have the rank the factors in the
xaxis and the eigenvalues in theyaxis.
Let’s do the scree plot to see the elbow point.
n <- dim(cor_mat)[1]
scree_tb <- tibble(x = 1:n,
y = sort(eigen(cor_mat)$value, decreasing = TRUE))
scree_plot <- scree_tb |>
ggplot(aes(x, y)) +
geom_point() +
geom_line() +
theme_minimal() +
scale_x_continuous(breaks = 1:n) +
ggtitle("Scree plot")
scree_plot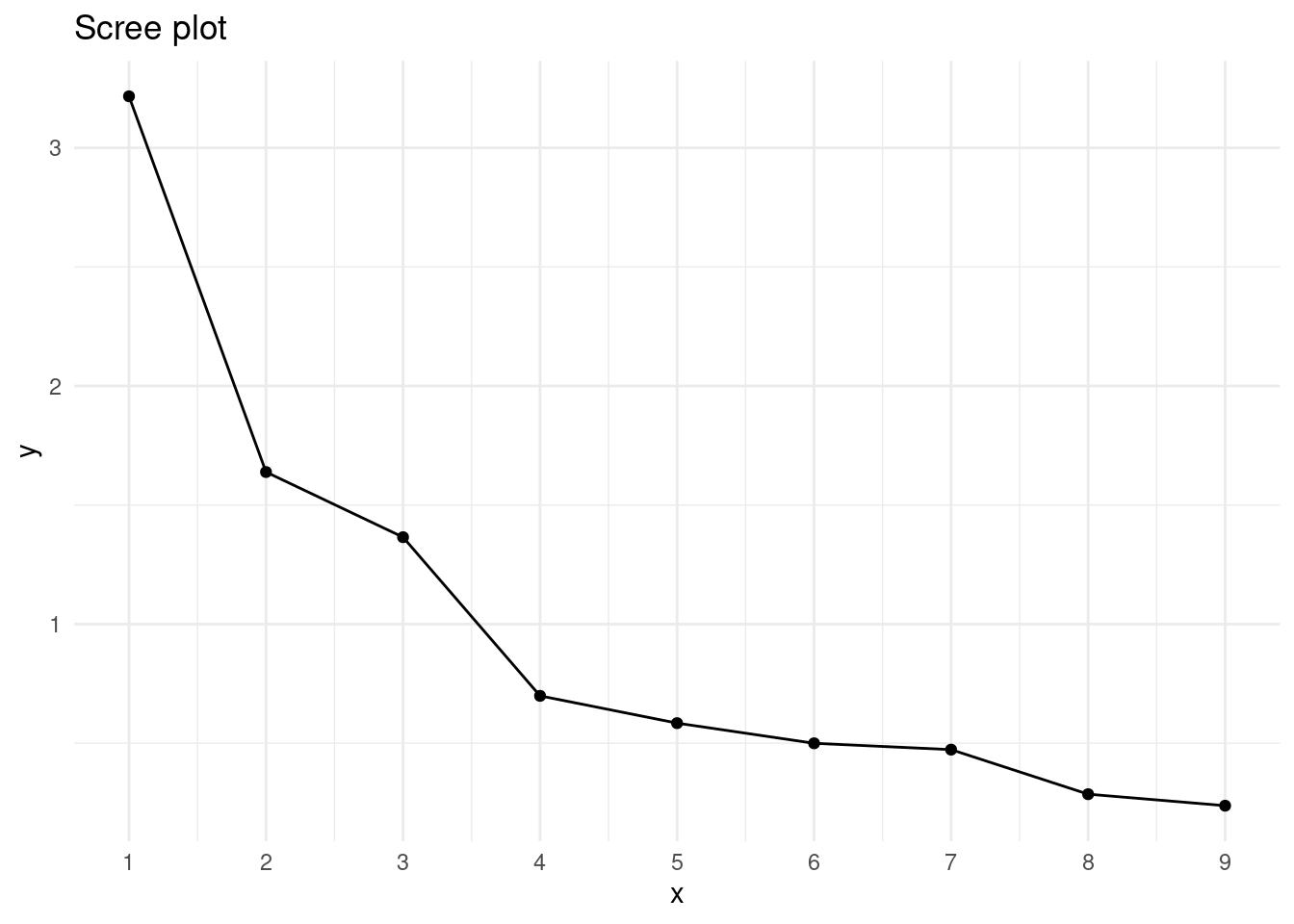
scree_plot +
geom_vline(xintercept = 4, color = "red", linetype = "dashed") +
annotate("text", 4.1, 2, label = "elbow point", color = "red", hjust = 0)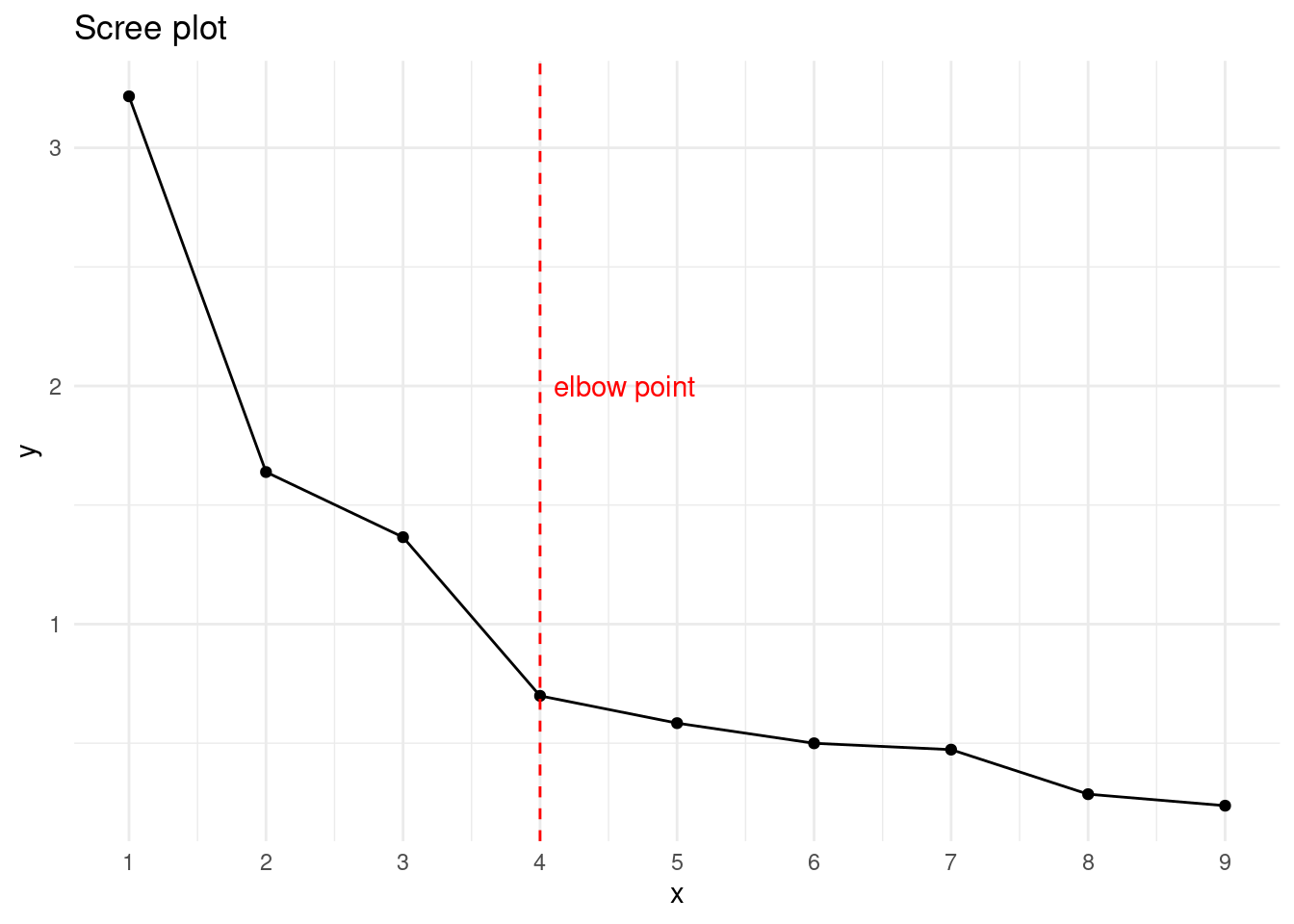
We observe that the elbow method occurs with four facgtor` and that we have three factors with eigenvalue greater than one. I have chosen to obtain a three-factor solution.
Factor Loadings and Rotations
Factor loedings \(f_{ij}\) are defined for each variable \(i\) and factor \(j\), and represent the correlation between both. A high value of \(f_{ij}\) means that a large amount of variability of variable \(i\) can be explained by factor \(j\).
Factor loadings of a model are unique up to a rotation. We prefer rotation methods that obtain factor loadings that allow relating one factor to each variable. Orthogonal rotations yield a set of uncorrelated factors. The most common orthogonal rotation methods are:
- Varimax maximizes variance of the squared loadings in each factor, so that each factor has only few variables with large loadings by the factor.
- Quartimax minimizes the number of factors needed to explain a variable.
- Equamax is a combination of varimax and quartimax.
Oblique rotations lead to a set of correlated factors. The most common oblique rotation methods are oblimin and promax.
In psych::fa, the type of rotation is passed with the rotate parameter.
For a factor analysis, we can split the variance of each variable in:
- Communality
h2: the proportion of the variance explained by factors. - Uniqueness
u2: the proportion of the variance not explained by factors, thus unique for the variable.
Interpreting the Results
We can perform the factor analysis using the psych::fa function. This function allows several methods to do the factor analysis specifying fm parameter. The main options available are:
fm = "minres"for the minimum residual solution (the default).fm = "ml"for the maximum likelihood solution.fm = "pa"for the principal axis solution.
Let’s do the factor analysis using the principal axis method and the varimax rotation. The other parameters are the correlation matrix and the number of factors nfactors = 3.
hz_factor <- fa(r = cor_mat, nfactors = 3, fm = "ml", rotate = "varimax")The output of the factor analysis performed above is:
hz_factor## Factor Analysis using method = ml
## Call: fa(r = cor_mat, nfactors = 3, rotate = "varimax", fm = "ml")
## Standardized loadings (pattern matrix) based upon correlation matrix
## ML1 ML3 ML2 h2 u2 com
## x1 0.28 0.62 0.15 0.49 0.51 1.5
## x2 0.10 0.49 -0.03 0.25 0.75 1.1
## x3 0.03 0.66 0.13 0.46 0.54 1.1
## x4 0.83 0.16 0.10 0.72 0.28 1.1
## x5 0.86 0.09 0.09 0.76 0.24 1.0
## x6 0.80 0.21 0.09 0.69 0.31 1.2
## x7 0.09 -0.07 0.70 0.50 0.50 1.1
## x8 0.05 0.16 0.71 0.53 0.47 1.1
## x9 0.13 0.41 0.52 0.46 0.54 2.0
##
## ML1 ML3 ML2
## SS loadings 2.18 1.34 1.33
## Proportion Var 0.24 0.15 0.15
## Cumulative Var 0.24 0.39 0.54
## Proportion Explained 0.45 0.28 0.27
## Cumulative Proportion 0.45 0.73 1.00
##
## Mean item complexity = 1.2
## Test of the hypothesis that 3 factors are sufficient.
##
## The degrees of freedom for the null model are 36 and the objective function was 3.05
## The degrees of freedom for the model are 12 and the objective function was 0.08
##
## The root mean square of the residuals (RMSR) is 0.02
## The df corrected root mean square of the residuals is 0.03
##
## Fit based upon off diagonal values = 1
## Measures of factor score adequacy
## ML1 ML3 ML2
## Correlation of (regression) scores with factors 0.93 0.81 0.84
## Multiple R square of scores with factors 0.87 0.66 0.70
## Minimum correlation of possible factor scores 0.74 0.33 0.40There is a lot of information here. Let’s see some relevant insights:
- The solution starts with the factor loadings for factors
ML1,ML3andML2. Factor loadings are the only values affected by the rotation. For each variable we obtain the communalitiesh2and uniquenessesu2. - Communalities are quite heterogeneous, ranging from 0.25 for
x2to 0.76 forx5. - The proportion of variance explained by each factor is presented in
Proportion Varand the cumulative variance inCumulative Var. From theCumulative Varwe learn that the model explains the 54% of total variance. This value is considered low for an exploratory factor analysis.
We can see better the pattern of factor loadings printing them with a cutoff value:
print(hz_factor$loadings, cutoff = 0.3)##
## Loadings:
## ML1 ML3 ML2
## x1 0.623
## x2 0.489
## x3 0.663
## x4 0.827
## x5 0.861
## x6 0.801
## x7 0.696
## x8 0.709
## x9 0.406 0.524
##
## ML1 ML3 ML2
## SS loadings 2.185 1.343 1.327
## Proportion Var 0.243 0.149 0.147
## Cumulative Var 0.243 0.392 0.539We can see that we can group variables x1 to x3 in a factor ML1, variables x4 to x6 in factor ML3 and variables x7 to x9 in factor ML2. For this dataset, this result is in correspondence with the meaning of the variables. ML1 would be realated with visual ability, ML3 with verbal ability and ML2 with mathematical ability.
Factor Scores
While factor loadings help to interpret the meaning of the factors, factor scores allow obtaining values of factors for each observation. The arguments of psych::factor.scores are the observations for which we want to compute the scores and the factor analysis model:
hz_scores <- factor.scores(HolzingerSwineford1939 |> select(x1:x9), hz_factor)The outcome of factor.scores is a list with matrices:
scoreswith the values of factor scores for each observation.weightswith the weights used to obtain the scores.r.scores, the correlation matrix of the scores.
Let’s see how factor scores look like:
hz_scores$scores |>
as_tibble() |>
slice(1:5) |>
kbl() |>
kable_styling(full_width = FALSE)| ML1 | ML3 | ML2 |
|---|---|---|
| 0.1222182 | -0.9011525 | 0.0418319 |
| -1.3415596 | 0.6430582 | 0.9741734 |
| -1.8725468 | -0.1570029 | -1.2605706 |
| -0.0706078 | 0.9977696 | -0.9695703 |
| -0.0528519 | -0.6743954 | 0.4465503 |
Let’s do a scatterplot of factors ML1 and ML3, highlighting the gender of each observation.
scores <- as_tibble(hz_scores$scores)
scores <- bind_cols(HolzingerSwineford1939 |> select(id, sex), scores) |>
mutate(sex = factor(sex))
scores |>
ggplot(aes(ML1, ML3, color = sex)) +
geom_point() +
theme_minimal() +
theme(legend.position = "bottom")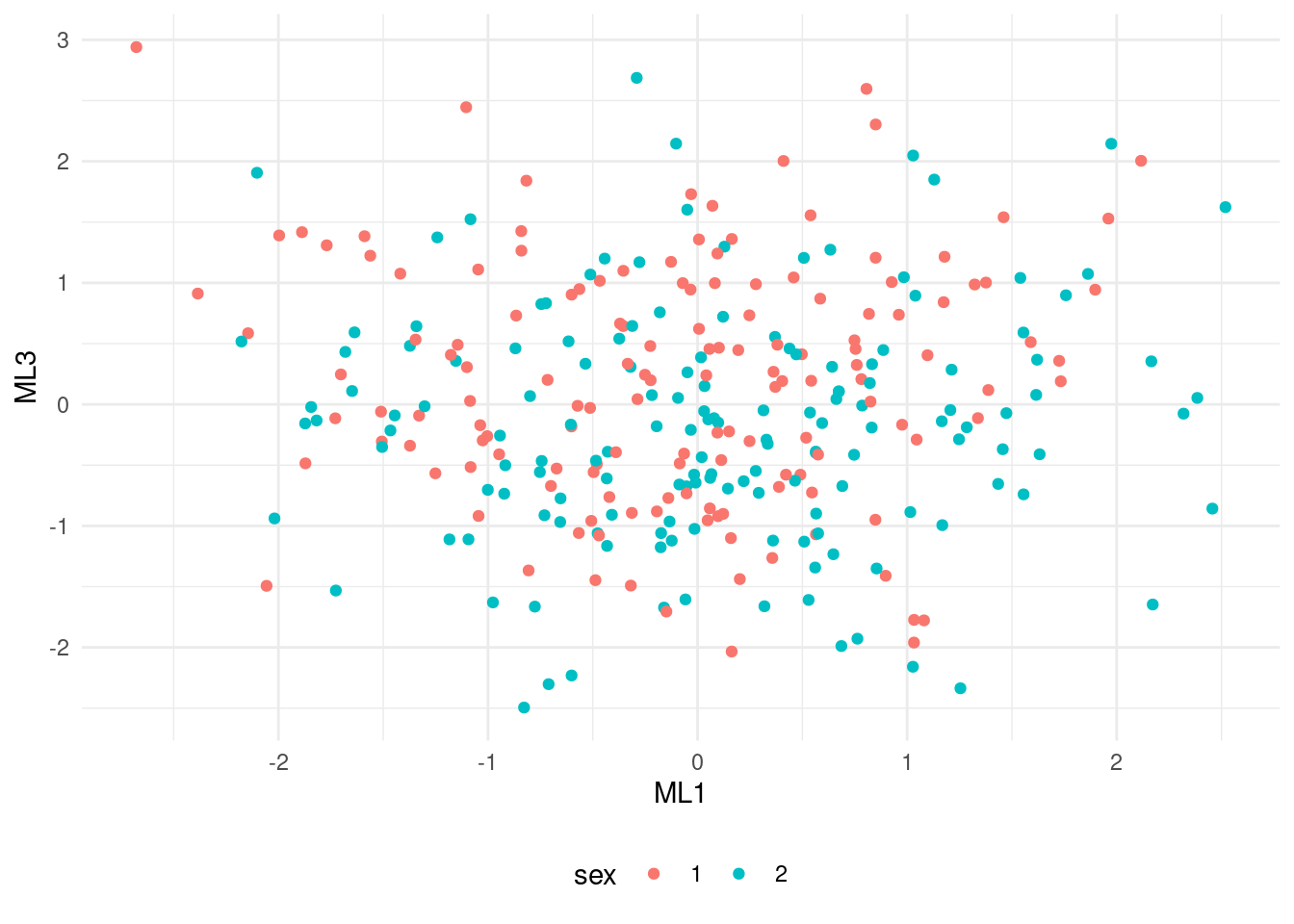
The plot show two uncorrelated factors, uniformly distributed among the two genders.
References
- Kaiser, H. F. (1974). An index of factor simplicity. Psychometrika, 39(1), 31-36.
- Statology (2019). A Guide to Bartlett’s Test of Sphericity. https://www.statology.org/bartletts-test-of-sphericity/
- Factor rotation methods (varimax, quartimax, oblimin, etc.) - what do the names mean and what do the methods do? https://stats.stackexchange.com/questions/185216/factor-rotation-methods-varimax-quartimax-oblimin-etc-what-do-the-names
Session Info
## R version 4.2.2 Patched (2022-11-10 r83330)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Linux Mint 21.1
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.10.0
## LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.10.0
##
## locale:
## [1] LC_CTYPE=es_ES.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=es_ES.UTF-8 LC_COLLATE=es_ES.UTF-8
## [5] LC_MONETARY=es_ES.UTF-8 LC_MESSAGES=es_ES.UTF-8
## [7] LC_PAPER=es_ES.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=es_ES.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] kableExtra_1.3.4 lavaan_0.6-14 psych_2.2.9 corrr_0.4.4
## [5] forcats_0.5.2 stringr_1.5.0 dplyr_1.0.10 purrr_1.0.1
## [9] readr_2.1.3 tidyr_1.3.0 tibble_3.1.8 ggplot2_3.4.0
## [13] tidyverse_1.3.2
##
## loaded via a namespace (and not attached):
## [1] nlme_3.1-162 fs_1.6.0 lubridate_1.9.1
## [4] webshot_0.5.4 httr_1.4.4 tools_4.2.2
## [7] backports_1.4.1 bslib_0.4.2 utf8_1.2.2
## [10] R6_2.5.1 DBI_1.1.3 colorspace_2.1-0
## [13] withr_2.5.0 tidyselect_1.2.0 mnormt_2.1.1
## [16] compiler_4.2.2 cli_3.6.0 rvest_1.0.3
## [19] TSP_1.2-2 xml2_1.3.3 labeling_0.4.2
## [22] bookdown_0.32 sass_0.4.5 scales_1.2.1
## [25] quadprog_1.5-8 systemfonts_1.0.4 digest_0.6.31
## [28] pbivnorm_0.6.0 rmarkdown_2.20 svglite_2.1.1
## [31] ca_0.71.1 pkgconfig_2.0.3 htmltools_0.5.4
## [34] dbplyr_2.3.0 fastmap_1.1.0 highr_0.10
## [37] rlang_1.0.6 readxl_1.4.1 rstudioapi_0.14
## [40] farver_2.1.1 jquerylib_0.1.4 generics_0.1.3
## [43] jsonlite_1.8.4 googlesheets4_1.0.1 magrittr_2.0.3
## [46] munsell_0.5.0 fansi_1.0.4 lifecycle_1.0.3
## [49] stringi_1.7.12 yaml_2.3.7 grid_4.2.2
## [52] parallel_4.2.2 crayon_1.5.2 lattice_0.20-45
## [55] haven_2.5.1 hms_1.1.2 knitr_1.42
## [58] pillar_1.8.1 codetools_0.2-19 stats4_4.2.2
## [61] reprex_2.0.2 glue_1.6.2 evaluate_0.20
## [64] blogdown_1.16 modelr_0.1.10 vctrs_0.5.2
## [67] tzdb_0.3.0 foreach_1.5.2 cellranger_1.1.0
## [70] gtable_0.3.1 assertthat_0.2.1 cachem_1.0.6
## [73] xfun_0.36 broom_1.0.3 googledrive_2.0.0
## [76] viridisLite_0.4.1 gargle_1.2.1 seriation_1.4.1
## [79] iterators_1.0.14 registry_0.5-1 timechange_0.2.0
## [82] ellipsis_0.3.2